Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 46(1); 2012 > Article
-
Original Article
Detection ofBRAF V600E Mutations in Papillary Thyroid Carcinomas by Peptide Nucleic Acid Clamp Real-Time PCR: A Comparison with Direct Sequencing - Dongjun Jeong,, Yujun Jeong,, Sungche Lee1, Hyeran Lee1, Wanju Lee2, Hyungjoo Kim, Doosan Park, Soyoung Park, Wenxia Mu, Hyun-Deuk Cho, Mee-Hye Oh, Sung Soo Lee3, Seung-Ha Yang, Chang-Jin Kim
-
Korean Journal of Pathology 2012;46(1):61-67.
DOI: https://doi.org/10.4132/KoreanJPathol.2012.46.1.61
Published online: February 23, 2012
Department of Pathology, Soonchunhyang University College of Medicine, Cheonan, Korea.
1Senior Undergraduate, Soonchunhyang University College of Medicine, Cheonan, Korea.
2College of Medical Sciences, Soonchunhyang University, Asan, Korea.
3Department of Preventive Medicine, Soonchunhyang University College of Medicine, Cheonan, Korea.
- Corresponding Author: Chang-Jin Kim, M.D. Department of Pathology, Soonchunhyang University College of Medicine, 366-1 Ssangyong-dong, Seobuk-gu, Cheonan 331-090, Korea. Tel: +82-41-570-2421, Fax: +82-41-575-2420, cjkim@sch.ac.kr
- *Dongjun Jeong and Yujun Jeong equally contributed to this work.
© 2012 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- Papillary thyroid carcinoma (PTC) of the thyroid is the most common endocrine malignancy. High prevalence of an activating point mutation of BRAF gene, BRAFV600E, has been reported in PTC. We assessed the efficiency of peptide nucleic acid clamp real-time polymerase chain reaction (PNAcqPCR) for the detection of BRAFV600E mutation in PTC in comparison with direct sequencing (DS).
-
Methods
- A total of 265 thyroid lesions including 200 PTCs, 5 follicular carcinomas, 60 benign lesions and 10 normal thyroid tissues were tested for BRAFV600E mutation by PNAcqPCR and DS.
-
Results
- The sensitivity and accuracy of the PNAcqPCR method were both higher than those of DS for the detection of the BRAFV600E mutation. In clinical samples, 89% of PTCs harbored the BRAFV600E mutation, whereas 5 follicular carcinomas, 50 benign lesions and 10 normal thyroid tissues lacked the mutation. The mutation was associated with aggressive clinical behaviors as extrathyroid invasion (p=0.015), lymph node metastasis (p=0.002) and multiple tumor numbers (p=0.016) with statistical significance.
-
Conclusions
- The PNAcqPCR method is efficiently applicable for the detection of the BRAFV600E mutation in PTCs in a clinical setting.
- Cell culture
- To validate the sensitivity and specificity of the commercial PNA Clamp BRAF mutation detection kit (Panagene Ltd., Daejeon, Korea), two human cancer cell lines (HeLa, Korea cell line bank No. 10002; BRAF wild type and COLO 205, Korea cell line bank No. 10222; BRAFV600E, T1799A, mutant type) were used as PNA clamping standards for real-time PCR. HeLa cells (BRAF wild type) were grown in Dulbecco's modified Eagle's medium (Hyclone, Thermo, Logan, UT, USA), while COLO 205 cells were grown in RPMI 1640, supplemented with 10% fetal bovine serum (Sigma, St. Louis, MO, USA). The cells were cultivated under standard conditions (humidified atmosphere, 5% CO2 in air, 37℃).
- Tumor samples
- A total of 205 cases of surgically resected thyroid malignant lesions (200 PTCs and 5 follicular carcinomas), 50 benign lesions (15 follicular adenomas, 10 cases of nodular hyperplasia and 10 cases of Hashimoto thyroiditis) and 10 normal thyroid tissues adjacent to nodular hyperplasia were included in this study. Eighty cases of papillary microcarcinoma (≤1 cm) were included to see if the BRAFV600E mutation occurs from early carcinogenesis. Informed consent was obtained from the patients for the collection of the tumor specimens, and the study protocol was approved by the Soonchunhyang University, College of Medicine Ethics Committee. The tissues were fixed in 10% neutral buffered formalin for 8-14 hours at room temperature for paraffin block.
- Preparation of genomic DNA
- Genomic DNAs were extracted from the cell lines using the High Pure PCR Template Preparation kit (Roche, Mannheim, Germany), according to the manufacturer's instructions, and the DNA was diluted to a concentration of 50 ng/µL for evaluation of the test. DNA from the COLO 205 cells, T1799A (V600E), was diluted with the DNA from the HeLa cells to give mutation/wild-type ratios of 1%, 5%, 10%, 20%, 50%, and 100%. The DNAs extracted from the cell lines were stored at -20℃ until use. The genomic DNAs from the formalin-fixed paraffin-embedded (FFPE) sections were extracted from 5×10 µm sections using the QIAmp DNA FFPE tissue kit (Qiagen, Valencia, CA, USA) as per the instructions.
- DNA sequencing
- The PCR reactions were performed in a final volume of 50 µL containing 1×PCR buffer, 200 µmol/L dNTPs, 200 nmol/L of each primer and 2.5 U of Taq polymerase (Solgent, Daejon, Korea). The forward primer was 5'-AAACTCTTCATAATGCTTGCTCTG-3'. The reverse primer was 5'-GGCCAAAAATTTAATCAGTGGA-3'. PCR was performed in a C1000 Thermal Cycler system (Bio-Rad, Hercules, CA, USA) with an initial denaturation temperature at 95℃ for 5 minutes followed by 40 cycles of 30 seconds at 95℃, 30 seconds at 62℃, and 1 minute at 72℃, and a final extension at 72℃ for 5 minutes. Amplicons of 231 bp were purified using the PCR purify kit according to the manufacturer's instructions (COSMO Co. Ltd., Seoul, Korea). Briefly, the amplicons were mixed with 5 volumes of PB buffer and the sample was loaded onto a column. After washing with NW buffer, the DNA was eluted with Tris buffer (10 mM Tris-Cl [pH 8.5]). DNA sequencing was performed with the PRISM 3100 DNA analyzer (Applied Biosystems, Carlsbad, CA, USA) using the ABI Prism BigDye Terminator ver. 3.1 (Applied Biosystems) with the forward primer.
- PNAcqPCR
- The BRAFV600E mutation was tested using the PNA Clamp BRAF mutation detection kit according to the manufacturer's instructions. Briefly, PCR was performed in a total volume of 20 µL that contained 50 ng of DNA, 13 µL of real-time SYBR Green PCR master mix and each of the primers and PNA probes for codon 600. The PCR control lacked a PNA probe and contained the wild type template. The PCR cycling conditions were at 94℃ for 5 minutes. followed by 40 cycles of four temperature steps (94℃ for 30 seconds, 70℃ for 20 seconds, 63℃ for 30 seconds and 72℃ for 30 seconds), and a final extension at 72℃ for 5 minutes. The PNA probe designed to hybridize completely to the wild-type BRAF allele securely inhibited the amplification of the wild-type BRAF allele, while the PNA/mutant-type allele hybrid was unstable due to base pair mismatch, which is why the extension by polymerase occurred. The threshold cycle (Ct) was automatically calculated from the PCR amplification plots where fluorescence was plotted against the number of cycles. Delta-Ct values were calculated as the Ct values of the samples minus those of the controls. The higher delta-Ct value showed that the mutant was efficiently amplified. A cut-off value of 2.0 was used for determining the presence of mutant DNA.
- Interpretation of the mutation data and statistical analysis
- For calculating sensitivity, specificity and accuracy of each detection method, true-positive (TP), true-negative (TN), false-positive (FP), and false-negative (FN) results were defined as follows. Detection of a BRAFV600E mutation in a tissue pathologically confirmed as PTC, in 200 cases, was considered as TP. Detection of a BRAFV600E mutation in a specimen pathologically confirmed as a normal, benign lesion or follicular carcinoma, in 75 cases, was categorized as FP. Failure to detect a BRAFV600E mutation in a normal, benign lesion or follicular carcinoma in 75 cases, was considered as TN, and failure to detect a BRAFV600E mutation in a PTC in 200 cases, was categorized as FN. Diagnostic values were calculated as follows: sensitivity=TP/(TP+FN)×100; specificity=TN/(TN+FP)×100; positive predictive value (PPV)=TP/(TP+FP)×100; negative predictive value=TN/(TN+FN)×100; accuracy=(TP+TN)/(TP+TN+FP+FN)×100. Analysis by the κ statistic was performed to evaluate the agreement between each method and histopathologic diagnosis (κ≥0.75, excellent; κ=0.40-0.75, fair or good; κ<0.40, poor). The χ2 test and Student's t-test were used to analyze the relationship between clinicopathologic features and BRAFV600E mutation in patients with PTC. A p-value of less than 0.05 was considered statistically significant.
MATERIALS AND METHODS
- Sensitivity and specificity of the PNAcqPCR compared to direct sequencing using cell lines for the detection of the BRAFV600E mutation
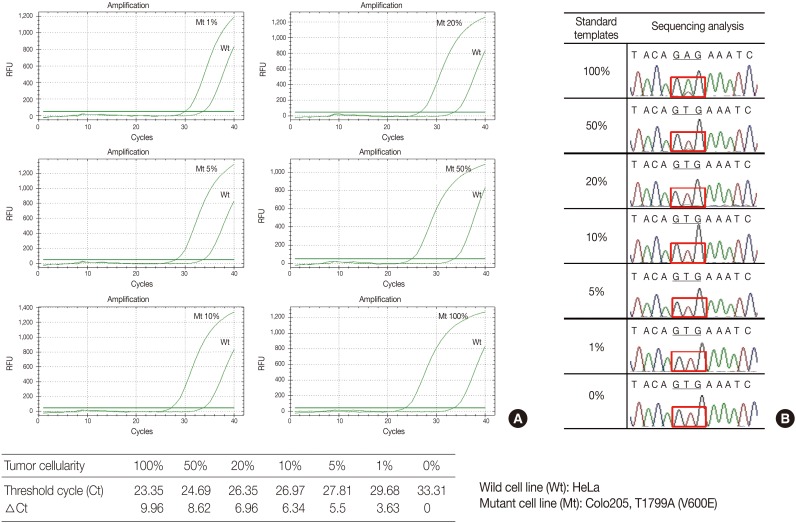
- The serially diluted mutant cells (COLO 205) in the wild type cell (HeLa) background were tested by PNAcqPCR in comparison to DS. The delta-Cts of the 1%, 5%, 10%, 20%, 50%, and 100% mutant were 3.63, 5.5, 6.34, 6.96, 8.62, and 9.96, respectively. The delta-Ct of the 1% mutant was larger than the cutoff value of 2.0 and the kit sensitively detected mutation in 1% of the mutants in the wild type background. For sequencing, the mutation was detected barely by manual reading in 50% of the mutants in the wild type background (Fig. 1).
- Detection of the BRAFV600E mutation in thyroid samples by PNAcqPCR compared to Direct Sequencing
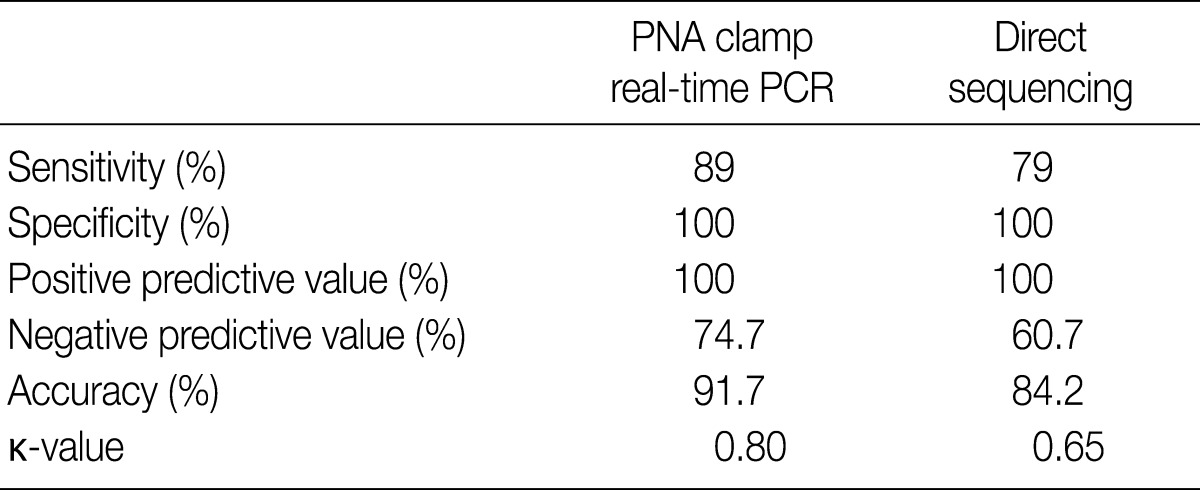
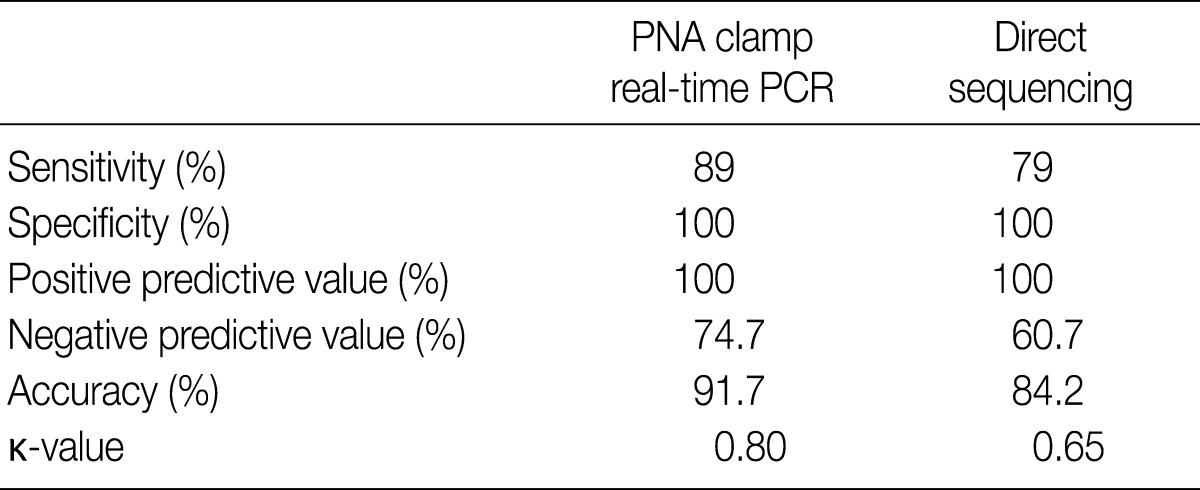
- Of the 200 PTC samples, 178 were positive (89%) by PNAcqPCR and 158 (79%) were positive by DS. All of the positive cases by DS were positive by PNAcqPCR as well. We failed to detect the BRAFV600E mutation either by PNAcqPCR or by DS in follicular carcinomas and benign thyroid lesions including follicular adenoma, nodular hyperplasia, Hashimoto thyroiditis and normal thyroid tissue. In 80 cases of papillary microcarcinoma, 71 cases (89%) were positive for BRAFV600E. The sensitivity to detect BRAFV600E by PNAcqPCR was 89% whereas it was 79% by DS. The specificity and PPV was 100% by the two methods. The accuracy to detect BRAFV600E was 91.7% by PNAcqPCR whereas it was 84.2% by DS. The Cohen's kappa coefficient of PNAcqPCR was 0.80, which is excellent, whereas that of DS was 0.65, which is fair or good (Table 1).
- Correlation between BRAFV600E mutation and clinicopathologic parameters in papillary thyroid carcinomas
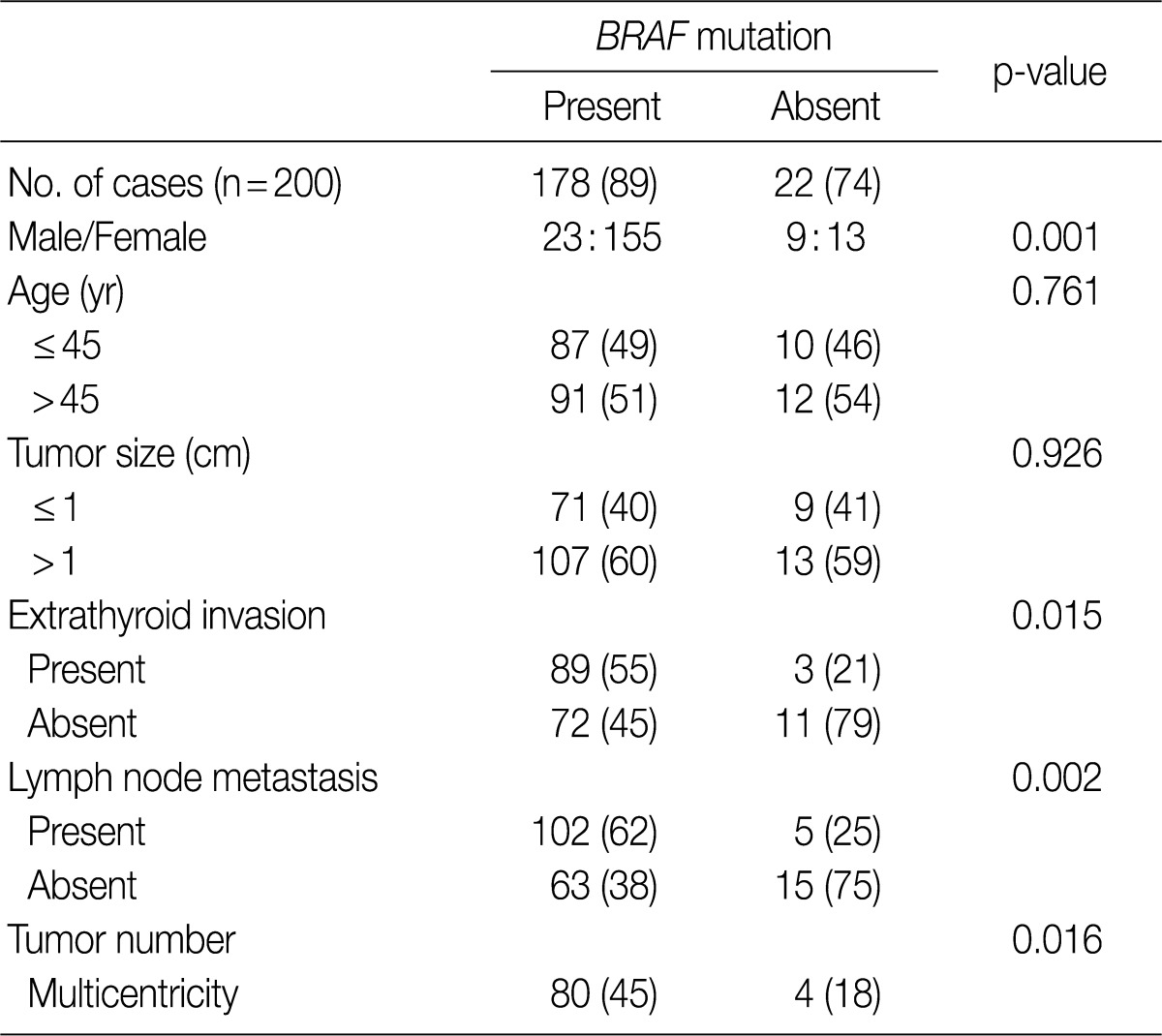
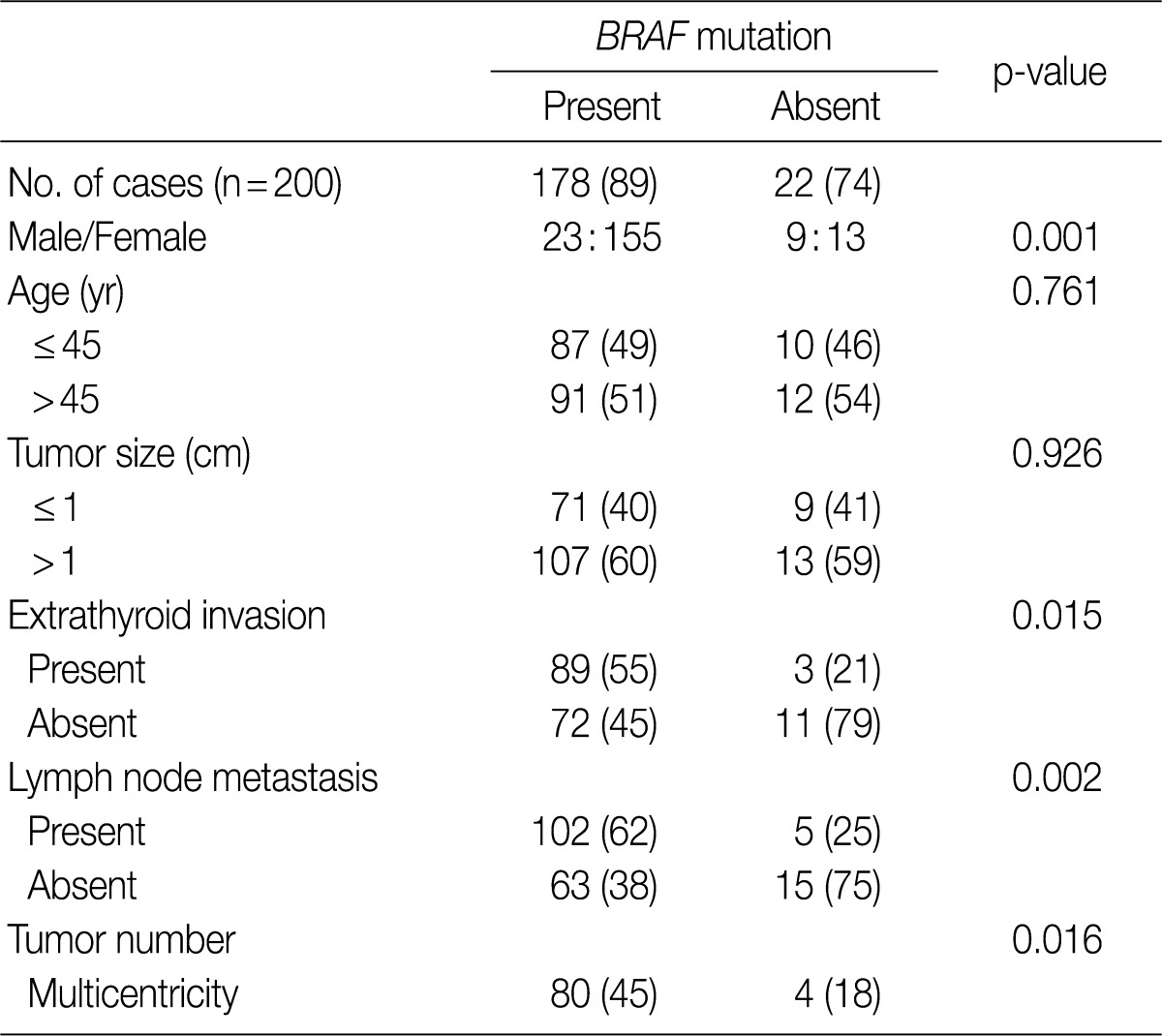
- We analyzed the association between the presence of the BRAFV600E mutation and various clinicopathologic parameters in patients with PTC, including sex, age, tumor size, extrathyroid invasion, lymph node metastasis and tumor multicentricity. As shown in Table 2, age and tumor size did not differ significantly in patients with or without the BRAFV600E mutation. The female patients had a higher prevalence of BRAFV600E which was statistically significant (p=0.001), compared to the male patients. The tumors with BRAFV600E had higher probabilities of extrathyroid extension (p=0.015). The tumors with nodal metastasis (p=0.002) and with multicentricity (p=0.016) had a statistically higher prevalence of BRAFV600E than that of non-metastasis and of single location (Table 2).
RESULTS
- PTC occurs due to oncogenic constitutive activation of the mitogen-activated protein kinase pathway by the BRAFV600E mutation.3 The prevalence of the BRAFV600E mutation in PTC is different according to the detection methods, ranging from 32% to 90%.3,4,10-18 In this study, the BRAFV600E mutation was detected in 89% (178/200) of PTC samples by the PNAcqPCR method, which is in concordance with the highest prevalence reported by Lee et al.18 In Korea, the BRAFV600E mutation has been reported to be present in 83-90% of PTCs,12,17,18 which is higher than what is reported (32-69%) in other countries.3,4,10,11,13-16 The reason behind higher prevalence of BRAFV600E in PTC in Korea compared to other countries is not clear. However, a recent epidemiologic study reported that high iodine intake may be a significant risk factor for the presence of the BRAFV600E mutation.22 Koreans usually consume sufficient iodine through the ingestion of seaweed products.23 Korean patients with thyroid diseases ingest considerably more iodine than healthy subjects, especially patients with thyroid cancer.24
- The PNAcqPCR detects the BRAFV600E mutation with high sensitivity. The COLO 205 cell line is a heterozygous mutant cell, and the sequencing chromatogram of 100% mutant cells demonstrated low wild type A peak. In order to test sensitivity to detect the BRAFV600E mutation, heterozygous mutant cells proportionally mixed with wild type cells may be used.25 As shown in this study with cell lines, the PNAcqPCR detected a 1% BRAF mutant in the background of the wild type with high sensitivity, though the gold standard DS detected a 50% mutant in the background of the wild type. The PNAcqPCR is a sensitive and specific method to detect point mutations.26,27 Several chemical features make PNA a superior PCR clamp for specific alleles. PNA cannot be used as a primer for polymerization, nor can it be used as a substrate for the exonuclease activities of Taq polymerase. In addition, the melting temperature (Tm) of a perfectly matched PNA-DNA duplex is higher than that of a DNA-DNA duplex of the same length or nucleotides, and a single mismatch destabilizes the PNA-DNA hybrids, causing a Tm shift.26 Therefore, a PNA probe can specifically block chain elongation on a perfectly matched wild template without interfering with these reactions on the mutant templates with mismatched bases.26,27 This enables the detection of mutants by real-time PCR rapidly and sensitively.
- DS is the "gold standard" which has been used to detect point mutations. This method, however, takes 2-3 days and has relatively low sensitivity, since samples must contain at least 20% mutant cells for successful mutant detection.19 In this study, DS failed to detect the BRAFV600E mutation in 20 samples that were positive for this mutation by PNAcqPCR. These samples were microcarcinomas that had diameters less than 1 mm with a few tumor cells.
- Comparing the efficiency between PNAcqPCR and DS for the detection of the BRAFV600E mutation, the sensitivity and accuracy of PNAcqPCR method were both higher than those of DS. However, the specificity and PPV of the two methods were both 100%. The PNAcqPCR method has several advantages over DS under clinical settings. First, it is more sensitive for the detection of the BRAFV600E mutation than DS. Second, it requires only simple real-time PCR equipment rather than an expensive sequencer. Third, the result can be obtained rapidly by this method. Fourth, as it uses a single tube reaction, cross contamination of the samples can be minimized.
- Although the BRAFV600E mutation is one of the most common genetic alterations in PTC, the exact biological characteristics, pathological features and clinical behaviors of tumors harboring BRAF mutations are still unclear. Lee et al.28 reported by meta-analysis including 1,168 patients from 12 studies that the BRAF mutation significantly correlated with histologic subtype (tall cell variant and conventional type), presence of extrathyroidal extension, and advanced clinical stage, but not with age, sex, race or tumor size. The association between the BRAFV600E mutation and lymph node metastasis was found to be of borderline significance (odds ratio, 1.5; 95% confidence interval, 0.992 to 2.268; p=0.055).
- In this study, the BRAFV600E mutation was associated with aggressive clinical behaviors including extrathyroid invasion (p=0.015), lymph node metastasis (p=0.002) and multiple tumor numbers (p=0.016) with statistical significance. This result is concordant with that of meta-analysis by Lee et al.28 It is noticeable that the BRAFV600E mutation occurs in 89% of papillary microcarcinomas. This shows that the BRAFV600E mutation plays a role in early carcinogenesis.
- Because of its high prevalence, and its association with aggressive clinical behaviors, BRAFV600E in PTC has become an attractive target for small molecular kinase inhibitors, which are currently in development. BAY43-9006 (sorafenib) and PLX4032 are novel signal transduction inhibitors that are intended to prevent tumor growth by blocking the RAS signaling pathway, and stop tumor angiogenesis by inhibiting the vascular endothelial growth factor receptor-2/platelet derived growth factor receptor-beta signaling cascade.29,30 Identification of the BRAFV600E activating point mutation by a rapid laboratory method could have significant clinical value, and maximize therapeutic efficacy by drugs used in PTC. The PNAcqPCR method could be a method of choice for the detection of BRAFV600E with sensitivity and rapidity. Also, it appears feasible to use both routine FFPE tissue samples and minute amount of cells obtained by fine needle aspiration.
- In summary, in this study, the BRAFV600E mutation was detected in 89% of PTCs, which is one of the highest levels of prevalence reported. This suggests that BRAFV600E provides a genetic marker differentiating papillary carcinomas from other types of thyroid carcinomas, and suggests that the BRAFV600E mutation can play a significant role in the carcinogenesis of papillary carcinoma. The BRAFV600E mutation was associated with aggressive tumor behaviors including extrathyroid invasion, multiplicity and regional lymph nodal metastasis. The PNAcqPCR method for the detection of BRAFV600E is sensitive, and is applicable in a clinical setting to maximize the therapeutic efficacy of small molecular kinase inhibitors.
DISCUSSION
Acknowledgments
Acknowledgments
- 1. Segev DL, Umbricht C, Zeiger MA. Molecular pathogenesis of thyroid cancer. Surg Oncol 2003; 12: 69-90. ArticlePubMed
- 2. Hay ID, Bergstralh EJ, Goellner JR, Ebersold JR, Grant CS. Predicting outcome in papillary thyroid carcinoma: development of a reliable prognostic scoring system in a cohort of 1779 patients surgically treated at one institution during 1940 through 1989. Surgery 1993; 114: 1050-1057. PubMed
- 3. Cohen Y, Xing M, Mambo E, et al. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst 2003; 95: 625-627. ArticlePubMed
- 4. Fukushima T, Suzuki S, Mashiko M, et al. BRAF mutations in papillary carcinomas of the thyroid. Oncogene 2003; 22: 6455-6457. ArticlePubMedPDF
- 5. Bonner TI, Kerby SB, Sutrave P, Gunnell MA, Mark G, Rapp UR. Structure and biological activity of human homologs of the raf/mil oncogene. Mol Cell Biol 1985; 5: 1400-1407. ArticlePubMedPMCPDF
- 6. Huleihel M, Goldsborough M, Cleveland J, Gunnell M, Bonner T, Rapp UR. Characterization of murine A-raf, a new oncogene related to the v-raf oncogene. Mol Cell Biol 1986; 6: 2655-2662. ArticlePubMedPMCPDF
- 7. Beck TW, Huleihel M, Gunnell M, Bonner TI, Rapp UR. The complete coding sequence of the human A-raf-1 oncogene and transforming activity of a human A-raf carrying retrovirus. Nucleic Acids Res 1987; 15: 595-609. ArticlePubMedPMC
- 8. Ikawa S, Fukui M, Ueyama Y, Tamaoki N, Yamamoto T, Toyoshima K. B-raf, a new member of the raf family, is activated by DNA rearrangement. Mol Cell Biol 1988; 8: 2651-2654. ArticlePubMedPMC
- 9. Kumar R, Angelini S, Czene K, et al. BRAF mutations in metastatic melanoma: a possible association with clinical outcome. Clin Cancer Res 2003; 9: 3362-3368. PubMed
- 10. Xing M, Westra WH, Tufano RP, et al. BRAF mutation predicts a poorer clinical prognosis for papillary thyroid cancer. J Clin Endocrinol Metab 2005; 90: 6373-6379. PubMed
- 11. Fugazzola L, Mannavola D, Cirello V, et al. BRAF mutations in an Italian cohort of thyroid cancers. Clin Endocrinol (Oxf) 2004; 61: 239-243. ArticlePubMed
- 12. Kim KH, Kang DW, Kim SH, Seong IO, Kang DY. Mutations of the BRAF gene in papillary thyroid carcinoma in a Korean population. Yonsei Med J 2004; 45: 818-821. ArticlePubMed
- 13. Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res 2003; 63: 1454-1457. PubMed
- 14. Rowe LR, Bentz BG, Bentz JS. Detection of BRAF V600E activating mutation in papillary thyroid carcinoma using PCR with allele-specific fluorescent probe melting curve analysis. J Clin Pathol 2007; 60: 1211-1215. ArticlePubMedPMC
- 15. Sapio MR, Posca D, Troncone G, et al. Detection of BRAF mutation in thyroid papillary carcinomas by mutant allele-specific PCR amplification (MASA). Eur J Endocrinol 2006; 154: 341-348. ArticlePubMed
- 16. Hayashida N, Namba H, Kumagai A, et al. A rapid and simple detection method for the BRAF(T1796A) mutation in fine-needle aspirated thyroid carcinoma cells. Thyroid 2004; 14: 910-915. ArticlePubMed
- 17. Kim SK, Song KH, Lim SD, et al. Clinical and pathological features and the BRAF(V600E) mutation in patients with papillary thyroid carcinoma with and without concurrent Hashimoto thyroiditis. Thyroid 2009; 19: 137-141. ArticlePubMed
- 18. Lee HJ, Choi J, Hwang TS, Shong YK, Hong SJ, Gong G. Detection of BRAF mutations in thyroid nodules by allele-specific PCR using a dual priming oligonucleotide system. Am J Clin Pathol 2010; 133: 802-808. ArticlePubMed
- 19. Lyon E, Wittwer CT. LightCycler technology in molecular diagnostics. J Mol Diagn 2009; 11: 93-101. ArticlePubMedPMC
- 20. Demers DB, Curry ET, Egholm M, Sozer AC. Enhanced PCR amplification of VNTR locus D1S80 using peptide nucleic acid (PNA). Nucleic Acids Res 1995; 23: 3050-3055. ArticlePubMedPMC
- 21. Jeong D, Jeong Y, Lee J, et al. Rapid and sensitive detection of KRAS mutation by peptide nucleic acid-based real-time PCR clamping: a comparison with direct sequencing between fresh tissue and formalin-fixed and paraffin embedded tissue of colorectal cancer. Korean J Pathol 2011; 45: 151-159. Article
- 22. Guan H, Ji M, Bao R, et al. Association of high iodine intake with the T1799A BRAF mutation in papillary thyroid cancer. J Clin Endocrinol Metab 2009; 94: 1612-1617. ArticlePubMedPDF
- 23. Kim JY, Moon SJ, Kim KR, Sohn CY, Oh JJ. Dietary iodine intake and urinary iodine excretion in normal Korean adults. Yonsei Med J 1998; 39: 355-362. ArticlePubMed
- 24. Kim JY, Kim KR. Dietary iodine intake and urinary iodine excretion in patients with thyroid diseases. Yonsei Med J 2000; 41: 22-28. ArticlePubMed
- 25. Ibrahem S, Seth R, O'Sullivan B, Fadhil W, Taniere P, Ilyas M. Comparative analysis of pyrosequencing and QMC-PCR in conjunction with high resolution melting for KRAS/BRAF mutation detection. Int J Exp Pathol 2010; 91: 500-505. ArticlePubMedPMC
- 26. Kyger EM, Krevolin MD, Powell MJ. Detection of the hereditary hemochromatosis gene mutation by real-time fluorescence polymerase chain reaction and peptide nucleic acid clamping. Anal Biochem 1998; 260: 142-148. ArticlePubMed
- 27. Thiede C, Bayerdorffer E, Blasczyk R, Wittig B, Neubauer A. Simple and sensitive detection of mutations in the ras proto-oncogenes using PNA-mediated PCR clamping. Nucleic Acids Res 1996; 24: 983-984. ArticlePubMedPMC
- 28. Lee JH, Lee ES, Kim YS. Clinicopathologic significance of BRAF V600E mutation in papillary carcinomas of the thyroid: a meta-analysis. Cancer 2007; 110: 38-46. ArticlePubMed
- 29. Ahmad T, Eisen T. Kinase inhibition with BAY 43-9006 in renal cell carcinoma. Clin Cancer Res 2004; 10(18 Pt 2):6388S-6392S. ArticlePDF
- 30. Sala E, Mologni L, Truffa S, Gaetano C, Bollag GE, Gambacorti-Passerini C. BRAF silencing by short hairpin RNA or chemical blockade by PLX4032 leads to different responses in melanoma and thyroid carcinoma cells. Mol Cancer Res 2008; 6: 751-759. ArticlePubMedPDF
REFERENCES
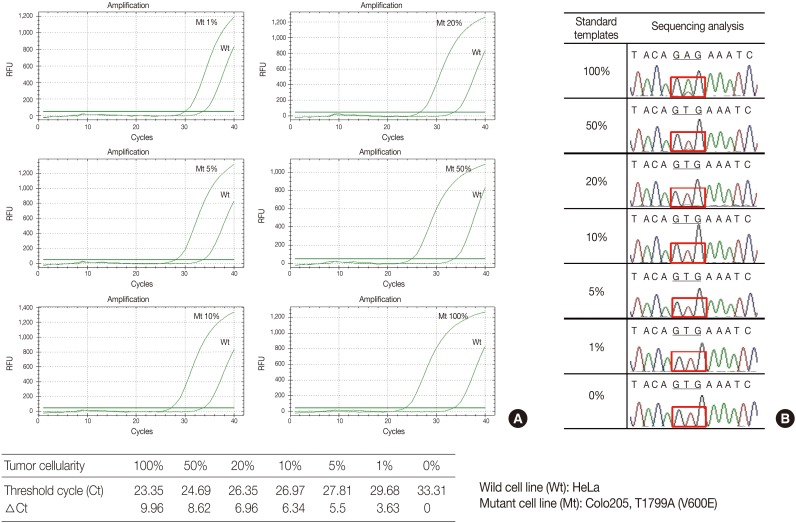
Figure & Data
References
Citations

- Diagnostic utilities of washout CYFRA 21-1 combined with washout thyroglobulin for metastatic lymph nodes in thyroid cancer: a prospective study
Joonseon Park, Solji An, Kwangsoon Kim, Jeong Soo Kim, Chan Kwon Jung, Ja Seong Bae
Scientific Reports.2024;[Epub] CrossRef - BRAFV600E Positivity-Dependent Effect of Age on Papillary Thyroid Cancer Recurrence Risk
Joonseon Park, Solji An, Kwangsoon Kim, Ja Seong Bae, Jeong Soo Kim
Cancers.2023; 15(22): 5395. CrossRef - TROP-2 and 5hmC expression in follicular-patterned thyroid neoplasm emphasizing tiny well-formed papillae
Jae Yeon Seok, Xuemo Fan
Annals of Diagnostic Pathology.2022; 57: 151903. CrossRef - Null Association between BRAF V600E Mutation and Tumor Recurrence in Patients with Papillary Thyroid Microcarcinoma in South Korea
Ji Yoon Kim, Kyoung Jin Kim, Jae Hyun Bae, Joo Hyung Kim, Nam Hoon Kim, Hee Young Kim, Hoon Yub Kim, Seung-Kuk Baek, Sin Gon Kim, Kwang Yoon Jung, Kyeong Jin Kim
International Journal of Thyroidology.2021; 14(2): 135. CrossRef - Evaluation of the expression levels of BRAFV600E mRNA in primary tumors of thyroid cancer using an ultrasensitive mutation assay
Tien Viet Tran, Kien Xuan Dang, Quynh Huong Pham, Ung Dinh Nguyen, Nhung Thi Trang Trinh, Luong Van Hoang, Son Anh Ho, Ba Van Nguyen, Duc Trong Nguyen, Dung Tuan Trinh, Dung Ngoc Tran, Arto Orpana, Ulf-Håkan Stenman, Jakob Stenman, Tho Huu Ho
BMC Cancer.2020;[Epub] CrossRef - The Current State of Molecular Testing in the BRAF-Mutated Melanoma Landscape
Irene Vanni, Enrica Teresa Tanda, Francesco Spagnolo, Virginia Andreotti, William Bruno, Paola Ghiorzo
Frontiers in Molecular Biosciences.2020;[Epub] CrossRef - Highly prevalent BRAF V600E and low-frequency TERT promoter mutations underlie papillary thyroid carcinoma in Koreans
Sue Youn Kim, Taeeun Kim, Kwangsoon Kim, Ja Seong Bae, Jeong Soo Kim, Chan Kwon Jung
Journal of Pathology and Translational Medicine.2020; 54(4): 310. CrossRef - Molecular pathology of thyroid tumours of follicular cells: a review of genetic alterations and their clinicopathological relevance
Giorgia Acquaviva, Michela Visani, Andrea Repaci, Kerry J Rhoden, Dario de Biase, Annalisa Pession, Tallini Giovanni
Histopathology.2018; 72(1): 6. CrossRef - Development of ultra-short PCR assay to reveal BRAF V600 mutation status in Thai colorectal cancer tissues
Nunthawut Chat-Uthai, Pichpisith Vejvisithsakul, Sutthirat Udommethaporn, Puttarakun Meesiri, Chetiya Danthanawanit, Yannawan Wongchai, Chinachote Teerapakpinyo, Shanop Shuangshoti, Naravat Poungvarin, John Souglakos
PLOS ONE.2018; 13(6): e0198795. CrossRef - T-blocker: a Simple and Robust Probe-Free Quantitative PCR Assay to Detect Somatic Mutations Down to 0.1% Frequency
Hanyoup Kim, Aaron E Ruby, Harini G Shandilya, Arvind K Virmani, Nandita Rahman, Christina M Strange, Jarkko Huuskonen
BioTechniques.2018; 65(4): 205. CrossRef - The usefulness of a novel fully automated PCR-based Idylla test for detection of the BRAF V600E mutation in thyroid tissue: comparison with PNA-clamping PCR, real-time PCR and pyrosequencing
Min-Kyung Yeo, Min-Kyu Jung, Su-Yel Lee, Yong-Moon Lee, Gang Min Hur, Jin-Man Kim
Journal of Clinical Pathology.2017; 70(3): 260. CrossRef - KRAS mutation analysis of washing fluid from endoscopic ultrasound-guided fine needle aspiration improves cytologic diagnosis of pancreatic ductal adenocarcinoma
Joo Kyung Park, Yoon Jung Lee, Jong Kyun Lee, Kyu Taek Lee, Yoon-La Choi, Kwang Hyuck Lee
Oncotarget.2017; 8(2): 3519. CrossRef - Application of Peptide Nucleic Acid-based Assays Toward Detection of Somatic Mosaicism
Christopher S Hong, Chunzhang Yang, Zhengping Zhuang
Molecular Therapy - Nucleic Acids.2016; 5: e314. CrossRef - Prediction Table and Nomogram as Tools for Diagnosis of Papillary Thyroid Carcinoma
Seo Ki Kim, Jun Ho Lee, Jung-Woo Woo, Inhye Park, Jun-Ho Choe, Jung-Han Kim, Jee Soo Kim
Medicine.2015; 94(21): e760. CrossRef - The association between BRAF V600E mutation and pathological features in PTC
Xin Liu, Kangkang Yan, Xuejun Lin, Longyu Zhao, Wenxiu An, Chunpeng Wang, Xiaodong Liu
European Archives of Oto-Rhino-Laryngology.2014; 271(11): 3041. CrossRef - Recurrent Thyroid Papillary Carcinoma in Children Under Ten Years Old: Report of Two Cases and Literature Review
Byeong-Joo Noh, Ji-Youn Sung, Youn-Wha Kim, Yong-Koo Park
Korean Journal of Pathology.2014; 48(4): 297. CrossRef - KRASMutation Detection in Non-small Cell Lung Cancer Using a Peptide Nucleic Acid-Mediated Polymerase Chain Reaction Clamping Method and Comparative Validation with Next-Generation Sequencing
Boram Lee, Boin Lee, Gangmin Han, Mi Jung Kwon, Joungho Han, Yoon-La Choi
Korean Journal of Pathology.2014; 48(2): 100. CrossRef - VE1 immunohistochemistry in pituitary adenomas is not associated with BRAF V600E mutation
Jan Sperveslage, Midea Gierke, David Capper, Jürgen Honegger, Bence Sipos, Rudi Beschorner, Jens Schittenhelm
Acta Neuropathologica.2013; 125(6): 911. CrossRef - BRAF Mutations in Iranian Patients with Papillary Thyroid Carcinoma
Nastran Ranjbari, Sara Almasi, Javad Mohammadi-asl, Fakher Rahim
Asian Pacific Journal of Cancer Prevention.2013; 14(4): 2521. CrossRef

Fig. 1
PNA, peptide nucleic acid; PCR, polymerase chain reaction.

 E-submission
E-submission
 PubReader
PubReader Cite this Article
Cite this Article