Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 50(5); 2016 > Article
-
Original Article
Long Non-coding RNA HOTAIR Expression in Diffuse Large B-Cell Lymphoma: In Relation to Polycomb Repressive Complex Pathway Proteins and H3K27 Trimethylation - Eun Ji Oh1,2,*, Soo Hee Kim1,3,4,*, Woo Ick Yang1, Young Hyeh Ko3, Sun Och Yoon,1
-
Journal of Pathology and Translational Medicine 2016;50(5):369-376.
DOI: https://doi.org/10.4132/jptm.2016.06.06
Published online: August 22, 2016
1Department of Pathology, Yonsei University College of Medicine, Seoul, Korea
2Department of Pathology, Seoul St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
3Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
4Anatomic Pathology Reference Lab, Seegene Medical Foundation, Seoul, Korea
- Corresponding Author Sun Och Yoon, MD, PhD Department of Pathology, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea Tel: +82-2-2228-1763 Fax: +82-2-362-0860 E-mail: soyoon@yuhs.ac; revita@naver.com
- *Eun Ji Oh and Soo Hee Kim contributed equally to this work.
© 2016 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- A long non-coding RNA hox transcript antisense intergenic RNA (HOTAIR) is involved in epigenetic regulation through chromatin remodeling by recruiting polycomb repressive complex 2 (PRC2) proteins (EZH2, SUZ12, and EED) that induce histone H3 trimethylation at lysine 27 (H3K27me3). Deregulation of c-MYC and interaction between c-MYC and EZH2 are well known in lymphomagenesis; however, little is known about the expression status of HOTAIR in diffuse large B-cell lymphomas (DLBCLs).
-
Methods
- The expression status of PRC2 (EZH2, SUZ12, and EED), H3K27me3, c-MYC, and BCL2 was analyzed using immunohistochemistry (n = 231), and HOTAIR was investigated by a quantification real-time polymerase chain reaction method (n = 164) in DLBCLs.
-
Results
- The present study confirmed the positive correlation among PRC2 proteins, H3K27me3, and c-MYC in DLBCLs. Expression level of HOTAIR was also positively correlated to EZH2 (p < .05, respectively). Between c-MYC and HOTAIR, and between c- MYC/BCL2 co-expression and HOTAIR, however, negative correlation was observed in DLBCLs (p < .05, respectively). High level of H3K27me3 was determined as an independent prognostic marker in poor overall survival (hazard ratio, 2.0; p = .023) of DLBCL patients. High expression of HOTAIR, however, was associated with favorable overall survival (p = .004) in the univariate analysis, but the impact was not significant in the multivariate analysis. The favorable outcome of DLBCL with HOTAIR high expression levels may be related to the negative correlation with c- MYC expression or c-MYC/BCL2 co-expression.
-
Conclusions
- HOTAIR expression could be one of possible mechanisms for inducing H3K27me3 via EZH2-related PRC2 activation, and induced H3K27me3 may be strongly related to aggressive DLBCLs which show poor patient outcome.
- Patients and clinical data
- A total of 231 cases of DLBCL treated with R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone) or R-CHOP–like (with or without radiotherapy or surgery) chemotherapy were selected for the study. Cases were retrieved from the archival files from the Department of Pathology, Severance Hospital, from 2005 to 2011. All cases were independently reviewed by two pathologists (S.O.Y. and S.H.K.) based on current World Health Organization criteria [9], and discordant cases were consulted to other expert hematopathologists. In HOTAIR expression analysis, 164 cases were selected from the above 231 cases and investigated after quality assessment of extracted RNA. Clinical data were obtained from medical records. All study protocols were performed according to the ethical guidelines of the ‘‘World Medical Association Declaration of Helsinki–Ethical Principles for Medical Research Involving Human Subjects.’’ This study was approved by the Institutional Review Board of Severance Hospital.
- Analysis for HOTAIR expression
- Formalin fixed paraffin embedded (FFPE) tissue sections were prepared and stained with hematoxylin and eosin, and then the tumor areas were confirmed and marked under the microscope. The marked areas mainly contained packed tumor cells, and the stromal component was less than 10% of the marked area. The unstained slides of FFPE tissues were prepared after dissecting FFPE tissue blocks at 10-mm thickness using a microtome, and the marked area was scraped using a scalpel blade. Generally, three slices of tissue section per case were used for RNA extraction. Total RNA was isolated using an RNeasy FFPE Kit (Qiagen, Hilden, Germany) according to the supplier’s instructions. Extracts of RNA were verified by measuring the ratios ofA260/A280 and A260/A230 with a ND-1000 NanoDrop spectrophotometer (NanoDrop, Wilmington, DE, USA). Reverse transcription was performed using a QuantiTect Reverse Transcription kit (Qiagen). The expression patterns of HOTAIR were assayed by relative quantification using expression of the house-keeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primers for HOTAIR and GAPDH were as follows: HOTAIR (forward, 5'-AGCCAGAGGAGGGAAGAGAG-3'; reverse, 5'-TCCCGTTCCCTAGATTTTCC-3') and GAPDH (forward, 5'-CAAATTCCATGGCACCGTCA-3'; reverse, 5'-ATCGCCCCACTTGATTTTGG-3'). Primers of HOTAIR were designed to detect all three transcript variants (transcript variant 1, 3, and 2). In brief, a 20 μL mixture containing 1.0 μL of cDNA, power SYBR Green PCR Master Mix (Applied Bio-systems, Carlsbad, CA, USA), 1.0 μL of 10 pmol/μL forward primer, 1.0 μL of 10 pmol/μL reverse primer, and 7.0 μL of tertiary distilled water was prepared. Amplification was performed using a Step One Plus Real-Time PCR instrument (Applied Biosystems) under the following conditions: denaturation at 94°C for 10 minutes, followed by 35 cycles of 94°C for 15 seconds, 55°C for 30 seconds, and 72°C for 30 seconds (fluorescence signal acquisition was performed at this phase). Immediately after amplification, melting curve analysis was performed for amplicon verification. All samples were analyzed in triplicate to confirm reproducibility. Using the Step One Plus Real-Time PCR System software ver. 2.1 (Applied Biosystems), the threshold cycle (Ct, beginning of the polymerase chain reaction exponential phase) value of amplified HOTAIR was normalized versus that of amplified GAPDH (2-dCt). After quality assessment of extracted RNA and expression analysis of the housekeeping gene GAPDH, 164 cases were finally selected for the analysis of HOTAIR expression. Normal palatine tonsil tissue was obtained from 10 cancer-free individuals and used as age-matched cancer-free controls. The cutoff value for high expression of HOTAIR (HOTAIRhigh) was determined at the uppermost value among those of normal control tonsil tissues.
- Tissue microarray preparation, immunohistochemistry, and analysis
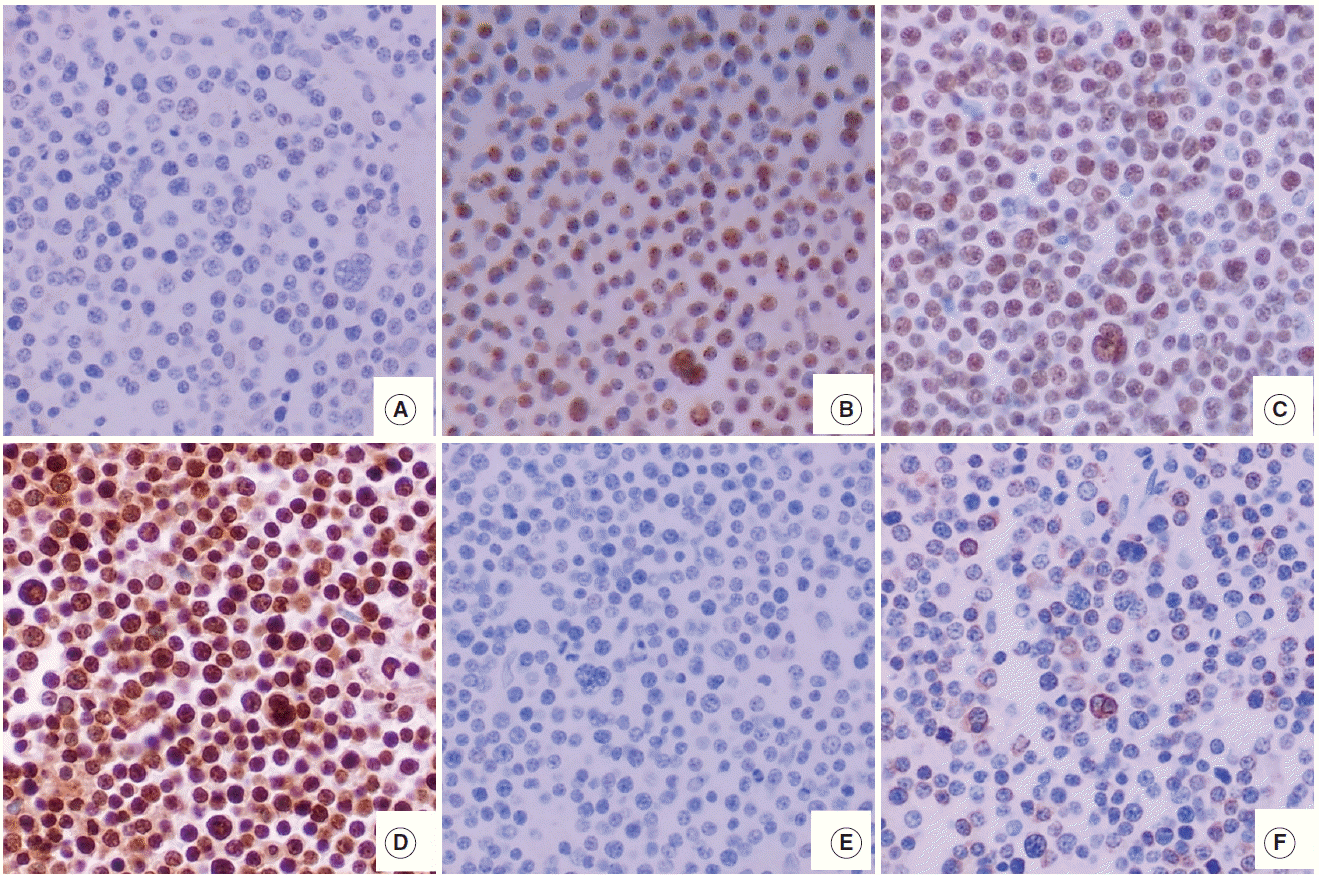
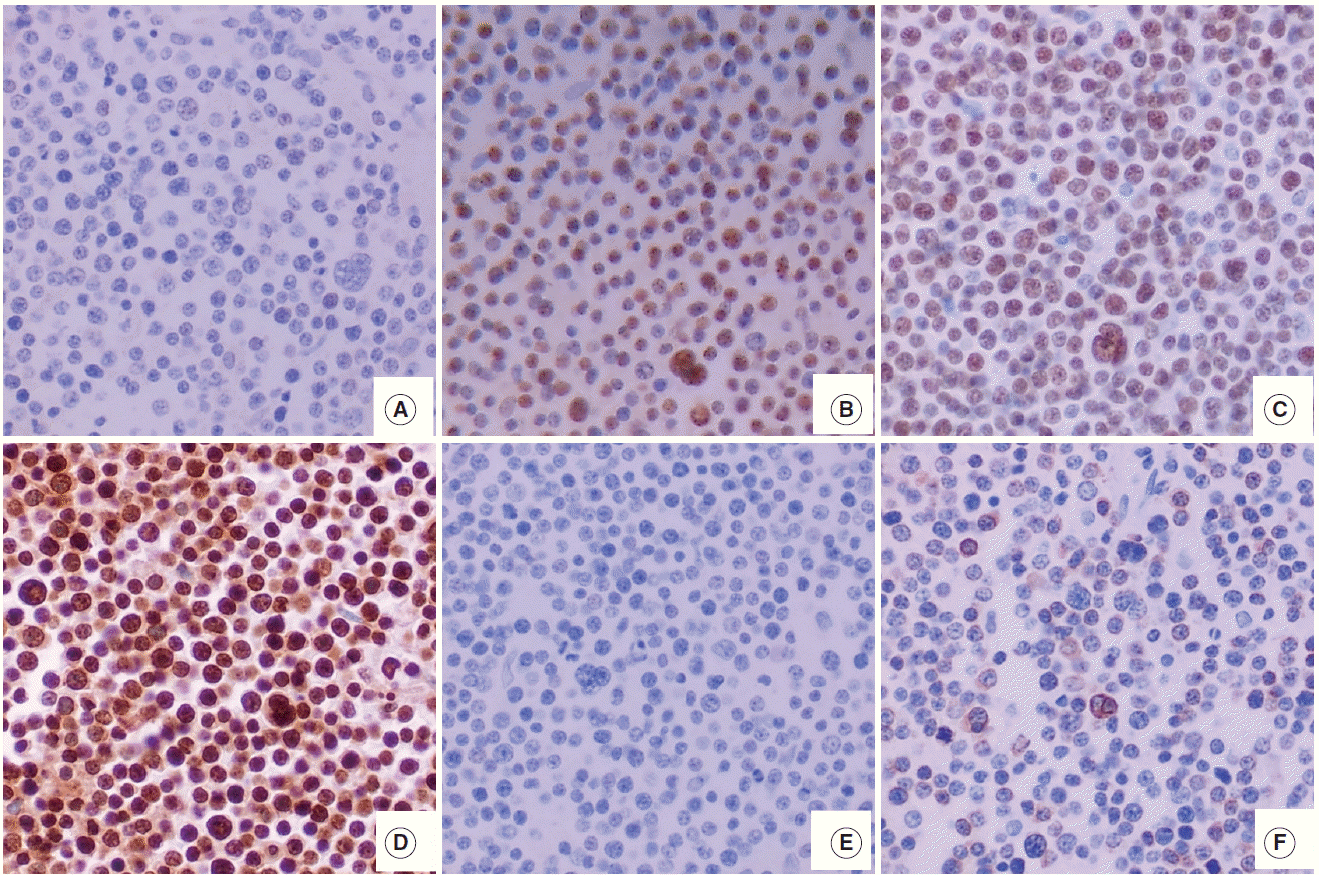
- The hematoxylin and eosin slides were reviewed and two representative core tissues of the tumor area were selected in each case. Core tissues (3 mm in diameter) were taken from the individual donor blocks and arranged in new recipient tissue microarray paraffin blocks using a trephine apparatus. Immunohistochemical staining and in situ hybridization were performed on 4-μm tissue microarray tissue sections. Immunohistochemistry of EZH2 (1:100, Invitrogen, Carlsbad, CA, USA), SUZ12 (1:50, Abcam, Cambridge, UK), EED (1:1,000, Abcam), H3K27me3 (1:100, clone C36B11, Cell Signaling Technology, Beverly, MA, USA) was performed using the Ventana BenchMark XT Autostainer (Ventana Medical Systems, Tucson, AZ, USA). Immunohistochemistry for c-MYC (1:50, clone Y69, Abcam), BCL2 (1:50, Novocastra, Newcastle, UK), CD10 (1:100, Novocastra), BCL6 (RTU, Novocastra), and MUM1 (1:200, Cell Marque, Rocklin, CA, USA) was performed using the LEICA BOND-III Autostainer (Leica Biosystems, Newcastle Upon Tyne, UK). For in situ hybridization for Epstein-Barr virus (EBV), the INFORM EBER probe (Ventana Medical Systems) was used with the Ventana BenchMark XT Autostainer (Ventana Medical Systems) and ISH iVIEW Blue Detection kit (Ventana Medical Systems).
- For EZH2, SUZ12, EED, and H3K27me3, staining intensity (0, no staining to weak; 1, moderate to strong) and proportion of positive tumor cell nuclei (0, <10%; 1, ≥10% and <75%; 2, ≥75%) were semiquantitatively graded as in the previous study [8]. Based on the intensity multiplied by proportion, the protein expression was scored as low (0), intermediate (1), or high level (2). For CD10, BCL6, and MUM1, the positive cutoff value was determined according to the Hans classification criteria [10] and was considered positive if ≥30% of the tumor cells showed nuclear immunoreactivity for BCL6 and MUM1, and if ≥30% of cells showed membranous reactivity for CD10. Determination of the germinal center B-like (GCB) or non-GCB phenotype was based on the Hans algorithm 10.
- For c-MYC and BCL2, staining intensity (0, no staining to weak; 1, moderate to strong) and areas of positive tumor cell nuclei by 10% increments (1 of <10% to 10 of 90%–100%) were semiquantitatively graded. The cutoff value for high expression was determined by log-rank tests (Mantel-Cox) for overall survival as in the previous report, and the threshold value was determined for c-MYC at a score ≥4 (≥40% of tumor cells with moderate to strong expression), and BCL2 at a score ≥7 (≥70% of tumor cells with moderate to strong expression) [11,12]. For EBV in situ hybridization, the threshold value was determined when ≥ 10% of the tumor cells showed moderate to strong nuclear expression.
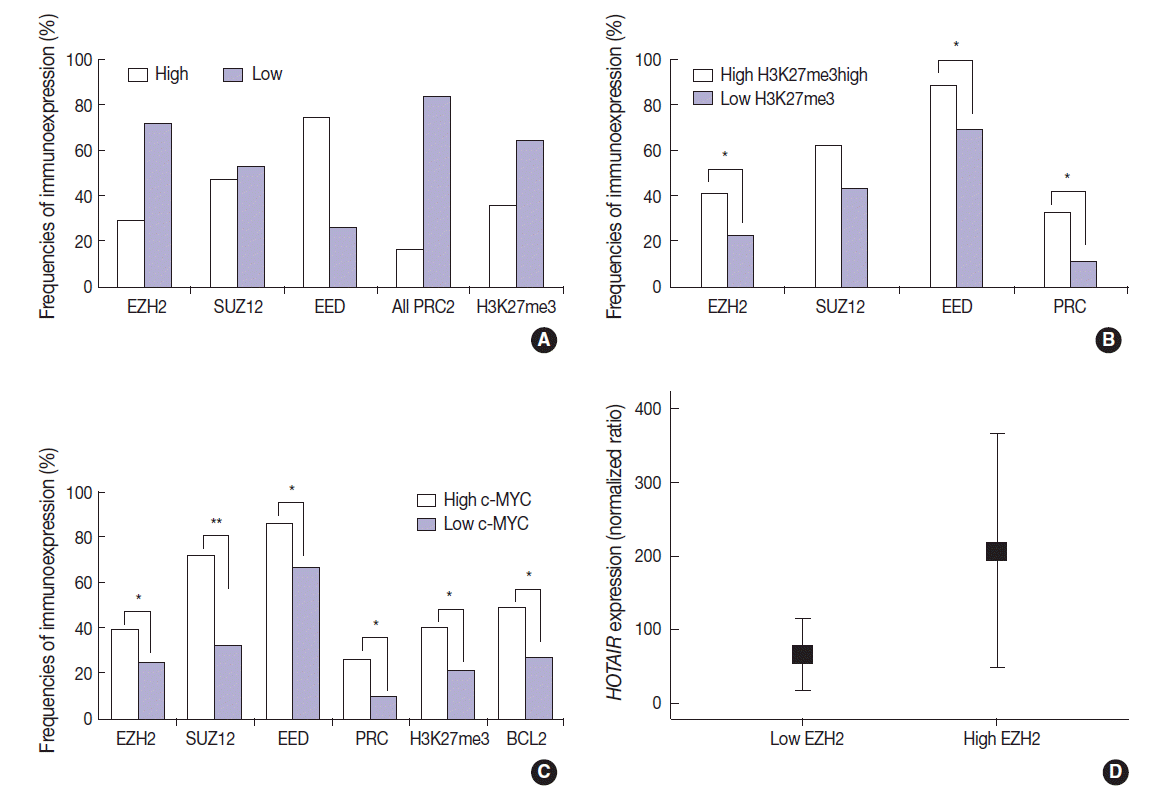
- The features of expression of polycomb repressive complex proteins (EZH2, SUZ12, and EED), H3K27me3, c-MYC, and BLC2 in HOTAIRhigh or HOTAIRlow cases are presented in Fig. 1.
- Statistical analysis
- The t test and chi-square test were used to analyze the differences between the variables examined. Overall survival times were measured from the date of lymphoma diagnosis to the date of death or last follow-up visit. Patient survival rates were determined using the Kaplan-Meier method, and the differences in survival rates were compared using the log-rank test. Multivariate analysis was performed using the Cox proportional hazards model. A two-sided p-value<.05 was considered to be statistically significant. When a two-sided p-value was ≤.05 and <.10, a trend toward statistical significance was considered. All statistical analyses were carried out using SPSS software ver. 20.0 for Windows (IBM Corp., Armonk, NY, USA).
MATERIALS AND METHODS
- The relations among PRC2 proteins, H3K27me3, c-MYC, BCL2, and HOTAIR
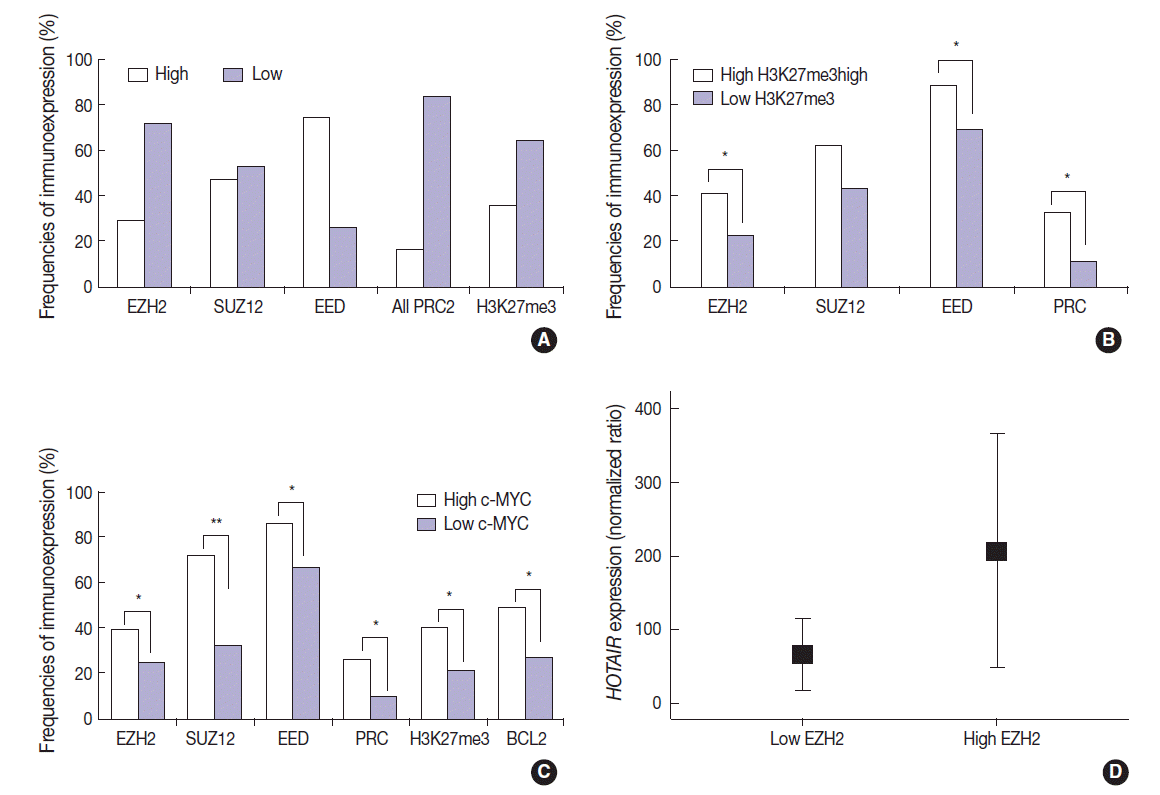
- The DLBCL cases frequently showed high expression of PRC2 proteins and H3K27me3 (28.9%, 47.1%, 74.3%, and 35.7% for EZH2high, SUZ12high, EEDhigh, and H3K27me3high, respectively). High expression of all PRC2 proteins (EZH2high/SUZ12high/EEDhigh) was noted in 16.1% of cases (Fig. 2A). The expression of H3K27me3 was positively correlated to PRC2 markers. When compared to cases of H3K27me3low, those of high H3K27me3 expression more frequently showed high expression of EZH2 (41.2% vs 22.2%, p=.003), SUZ12 (62.5% vs 43.0%, p=.053), EED (88.6% vs 69.7%, p=.026), and all PRC2 (EZH2high/SUZ12high/EEDhigh) (32.3% vs 11.3%, p=.005). These results are summarized in Fig. 2B.
- The expression of c-MYC was positively correlated to PRC2 markers and H3K27me3. When compared to cases of c-MYClow, those of high c-MYC expression more frequently showed high expression of EZH2 (39.1% vs 24.6%, p=.036), SUZ12 (72.2% vs 32.1%, p<.001), EED (85.7% vs 66.7%, p=.011), all PRC2 (EZH2high/SUZ12high/EEDhigh) (26.4% vs 9.5%, p=.009), and H3K27me3 (40.6% vs 21.2%; p=.004). BCL2 was positively correlated with c-MYC (49.3% vs 26.8%; p=.002). These results are summarized in Fig. 2C. BCL2 expression showed no significant correlation with the tested PRC2-related markers.
- The expression level of HOTAIR was significantly higher in cases of EZH2high than those of EZH2low (mean ratio value, 207 vs 66; p=.027) (Fig. 2D). For other markers, no statistical significance was observed according to HOTAIR expression level.
- HOTAIR expression in in relation to clinicopathological variables of DLBCL
- In the present study, 23.8% of cases (39/164) showed high HOTAIR expression levels (HOTAIRhigh). In the correlation analysis between HOTAIRhigh and clinicopathological characteristics of DLBCL (Table 1), high expression of c-MYC protein was less frequent in HOTAIRhigh than in HOTAIRlow (p=.037). The rate of co-expression of c-MYC and BCL2 was significantly lower in HOTAIRhigh than in HOTAIRlow (p=.015). Other clinicopathological factors including EBV (p=.660) and Hans classification (p=.746) showed no significant correlation with HOTAIR expression (Table 1).
- Clinical significance of RNA and protein expression
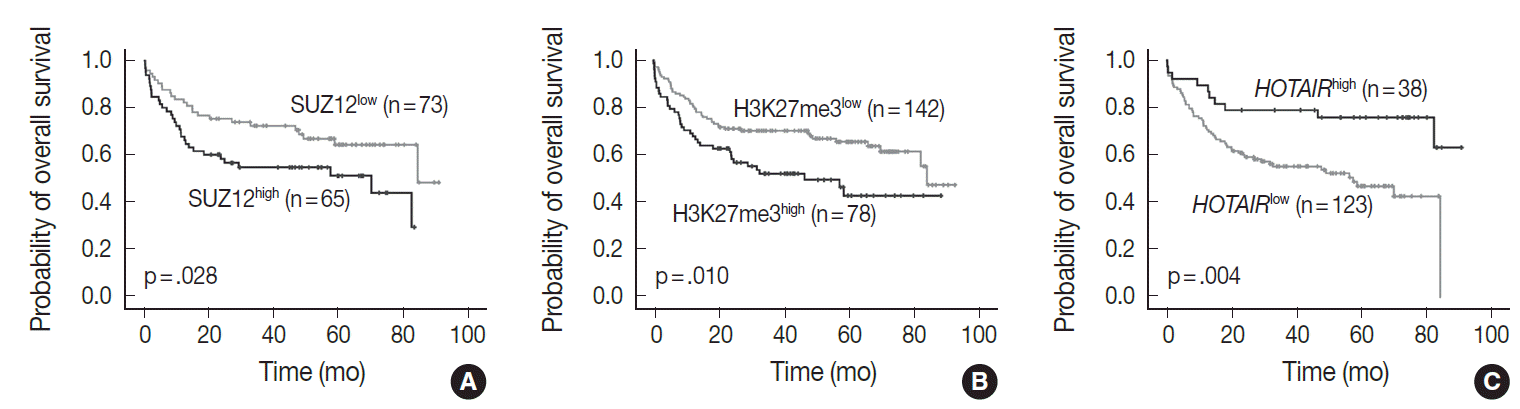
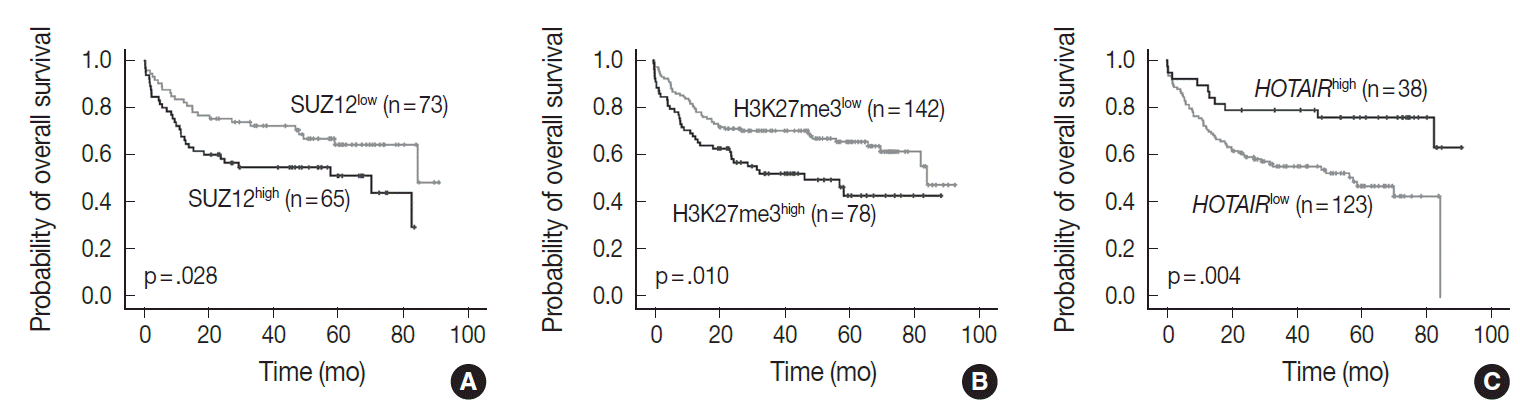
- In univariate analysis for overall survival of DLBCL patients (Table 2), the known prognostic factors including age >60 (p<.001), Eastern Cooperative Oncology Group performance status ≥2 (p<.001), elevated lactate dehydrogenase (p<.001), extranodal sites ≥2 (p=.001), Ann-Arbor stage III–IV (p=.003), and International Prognostic Index (IPI) risk ≥3 (p<.001) showed significant association with reduced overall survival rate. Expression of c-MYC and co-expression of c-MYC/BCL2 were also related to inferior overall survival rate (p=.032 and p=.006, respectively). Among the PRC pathway markers (EZH2, SUZ12, EED, and H3K27me3), SUZ12high and H3K27me3high were related to inferior overall survival rate (p=.028 and p=.010, respectively) (Fig. 3A, B). HOTAIRhigh, however, was significantly related to superior overall survival rate than HOTAIRlow (p=.004) (Fig. 3C).
- In multivariate analysis (Table 2), H3K27me3high revealed an independent effect on poor overall survival (hazard ratio, 2.0; p=.023). IPI risk ≥3 was still determined as an independent prognostic factor (hazard ratio, 3.5; p<.001), while HOTAIRhigh showed a tendency to be related to improved survival with marginal significance (hazard ratio, 0.5; p=.086).
RESULTS
- In our previous study, high level of global H3K27me3 was found to be a negative prognostic indicator in patients with DLBCL [8]. This subsequent study aimed to explore the role of HOTAIR, possibly functioning via induction of H3K27me3. We found the positive correlation between PRC2 proteins, global H3K27me3 levels, and c-MYC in DLBCL as expected.
- In the present study, the positive correlation among PRC2 proteins (EZH2, SUZ12, and EED), global H3K27me3 levels, and c-MYC was also confirmed in DLBCL as expected. In addition, HOTAIR expression was related to EZH2 expression. These findings could support that the lncRNA HOTAIR may be involved in inducing H3K27me3 through recruiting polycomb repressive complex, the methyltransferase EZH2 and core accessory proteins, SUZ12 and EED.
- Based on many other studies, interaction between c-MYC and PRC2 (EZH2, SUZ12, and EED) is well known in the tumorigenesis of various cancer types including lymphomas. Protein c-MYC interacts with EZH2 as well as SUZ12/EED, and they induce the histone modification of H3K27me3 on the promoter of target genes, which then represses gene expression. Many other studies have shown that EZH2 and c-MYC activate each other [13-15]. From the present findings, it could be also suggested that c-MYC may be the possible mechanism for inducing H3K27me3 via PRC2-related pathways in DLBCLs.
- In other studies of solid tumors originating from various organs, high expression of HOTAIR showed a close association with poor prognosis [5]. In the present study, however, high levels of HOTAIR expression showed an association with good prognosis in DLBCL, although the impact was not significant in the multivariate analysis. There are no comparable reports to support the present findings because this is the first study for the expression status of HOTAIR in hematologic malignancies.
- Although interaction between c-MYC and HOTAIR has not been established in hematologic malignancies, a close relationship between them may be plausible when considering the close association of PRC2 and HOTAIR, and PRC2 and c-MYC. However, expression of c-MYC showed a negative correlation with HOTAIR in the present study. One of the possible reason is autoregulation of c-MYC reported in one study; c-MYC represses itself via forming autoregulatory loops with EZH [2,13] therefore, HOTAIR might be involved in that process via EZH2. Whether HOTAIR expression is directly linked to the suppression of c-MYC or H3K27me3 should be investigated in further functional studies. In the HOTAIR-related suppression of c-MYC, PRC2-associated histone modification (H3K27me3) might be induced in the promoter region of c-MYC gene. For confirmation of this possible modulation, further study should be followed.
- The favorable outcome of DLBCLs with high HOTAIR expression might be associated with the negative correlation with c-MYC and/or BCL2. Recent evidence has shown that co-expression of c-MYC and BCL2 proteins is associated with poor prognosis in DLBCL patients regardless of gene signature [11,12]. This was also observed in the present study in the univariate survival analysis. When the factor of HOTAIR expression was added in the multivariate analysis; however, the prognostic effect of coexpression of c-MYC and BCL2 became weak, and only the factor of high H3K27me3 level was important as well as the IPI risk score. H3K27me3, the chromatin modification status induced via changes of HOTAIR, c-MYC, or other various known and unknown mechanisms, seems to be the most important factor in determining the fate of disease aggressiveness of DLBCL. Though little is known about the key role of HOTAIR in the malignant lymphoma, HOTAIR might be involved in regulation of various genes in the lymphomagenesis. Further study should follow to determine the explicit mechanism among the sophisticated modulatory networks of c-MYC, PRC2, H3K27me3, and HOTAIR.
- Recent evidence has shown that inhibition of EZH2 methyltransferase activity provides a potential target therapy for EZH2-deregulated lymphomas. In addition, the pharmacological inhibition of EZH2 activity results in a decrease of global H3K27me3 levels and a reactivation of silenced PRC2 gene targets, and it inhibits growth of DLBCL cells [16,17]. From these overall findings as well as the present findings, HOTAIR may also be used in a target therapy by modulating the c-MYC–EZH2/PRC2 loop in a subset of DLBCLs with a high level of H3K27me3.
- In conclusion, we found frequent expression of PRC2 proteins and H3K27me3 and positive correlation between these proteins and c-MYC in DLBCLs. High expression of H3K27me3 was determined as an independent predictor of poor prognosis, however, HOTAIR was associated with favorable overall survival, which can be partly explained by negative correlation with c-MYC. LncRNA HOTAIR expression could be one of the possible mechanisms to be involved in aggressive behavior of DLBCL via induction of H3K27me3 and EZH2-related PRC2 activation.
DISCUSSION
Acknowledgments
| Variable | No. (%) (n = 164) |
HOTAIR |
p-value | |
|---|---|---|---|---|
| HOTAIRlow | HOTAIRhigh | |||
| Sex | ||||
| Male | 98 (59.8) | 73 (58.4) | 25 (64.1) | .526 |
| Female | 66 (40.2) | 52 (41.6) | 14 (35.9) | |
| Age (yr) | ||||
| ≤ 60 | 94 (57.3) | 69 (55.2) | 25 (64.1) | .326 |
| > 60 | 70 (42.7) | 56 (44.8) | 14 (35.9) | |
| ECOG performance statusa | ||||
| < 2 | 132 (80.5) | 96 (76.8) | 36 (92.3) | .033 |
| ≥ 2 | 32 (19.5) | 29 (23.2) | 3 (7.7) | |
| Ann-Arbor stagea | ||||
| I–II | 89 (54.6) | 63 (50.8) | 26 (66.7) | .083 |
| III–IV | 74 (45.4) | 61 (49.2) | 13 (33.3) | |
| Extranodal sitesa | ||||
| < 2 | 108 (66.3) | 81 (65.3) | 27 (69.2) | .653 |
| ≥ 2 | 55 (33.7) | 43 (34.7) | 12 (30.8) | |
| Lactate dehydrogenasea | ||||
| Normal | 90 (55.2) | 65 (52.4) | 25 (64.1) | .201 |
| Elevated | 73 (44.8) | 59 (47.6) | 14 (35.9) | |
| IPI risk groupa | 79 (63.7) | 28 (71.8) | .354 | |
| 0–2 | 107 (65.6) | 45 (36.3) | 11 (28.2) | |
| 3–5 | 56 (34.4) | |||
| Hans classificationa | ||||
| GCB | 44 (28.0) | 32 (27.1) | 12 (30.8) | .660 |
| Non-GCB | 113 (72.0) | 86 (72.9) | 27 (69.2) | |
| EBV | ||||
| Negative | 132 (88.0) | 98 (87.5) | 34 (89.5) | .746 |
| Positive | 18 (12.0) | 14 (12.5) | 4 (10.5) | |
| BCL2a | ||||
| Negative expression | 84 (58.7) | 58 (54.7) | 26 (70.3) | .098 |
| Positive expression | 59 (41.3) | 48 (45.3) | 11 (29.7) | |
| c-MYCa | ||||
| Negative expression | 92 (62.2) | 63 (57.3) | 29 (76.3) | .037 |
| Positive expression | 56 (32.8) | 47 (42.7) | 9 (23.7) | |
| c-MYC/BCL2a | ||||
| No co-expression | 110 (77.5) | 76 (72.4) | 34 (91.9) | .015 |
| Co-expression | 32 (22.5) | 29 (27.6) | 3 (8.1) | |
HR, hazard ratio; CI, confidence interval; ECOG PS, Eastern Cooperative Oncology Group performance status; IPI, International Prognostic Index; EBV, Epstein-Barr virus; EZH2, enhancer of zeste homolog 2; SUZ12, suppressor of zeste 12 homolog; EED, embryonic ectoderm development; HOTAIR, hox transcript antisense intergenic RNA.
- 1. Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene 2012; 31: 4577-87. ArticlePubMedPMCPDF
- 2. Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 2013; 12: 433-46. ArticlePubMedPDF
- 3. Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071-6. ArticlePubMedPMCPDF
- 4. Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol 2013; 34: 613-20. ArticlePubMedPDF
- 5. Zhang S, Chen S, Yang G, et al. Long noncoding RNA HOTAIR as an independent prognostic marker in cancer: a meta-analysis. PLoS One 2014; 9: e105538.ArticlePubMedPMC
- 6. Martin-Perez D, Piris MA, Sanchez-Beato M. Polycomb proteins in hematologic malignancies. Blood 2010; 116: 5465-75. ArticlePubMedPDF
- 7. Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 2009; 9: 773-84. ArticlePubMedPDF
- 8. Oh EJ, Yang WI, Cheong JW, Choi SE, Yoon SO. Diffuse large B-cell lymphoma with histone H3 trimethylation at lysine 27: another poor prognostic phenotype independent of c-Myc/Bcl2 coexpression. Hum Pathol 2014; 45: 2043-50. ArticlePubMed
- 9. Swerdlow SH, Campo E, Harris NL, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon: IARC Press, 2008.
- 10. Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004; 103: 275-82. ArticlePubMed
- 11. Johnson NA, Slack GW, Savage KJ, et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol 2012; 30: 3452-9. PubMedPMC
- 12. Hu S, Xu-Monette ZY, Tzankov A, et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood 2013; 121: 4021-31. ArticlePubMedPMCPDF
- 13. Benetatos L, Vartholomatos G, Hatzimichael E. Polycomb group proteins and MYC: the cancer connection. Cell Mol Life Sci 2014; 71: 257-69. ArticlePubMedPDF
- 14. Sander S, Bullinger L, Klapproth K, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood 2008; 112: 4202-12. ArticlePubMedPDF
- 15. Neri F, Zippo A, Krepelova A, Cherubini A, Rocchigiani M, Oliviero S. Myc regulates the transcription of the PRC2 gene to control the expression of developmental genes in embryonic stem cells. Mol Cell Biol 2012; 32: 840-51. ArticlePubMedPMCPDF
- 16. McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012; 492: 108-12. ArticlePubMedPDF
- 17. Qi W, Chan H, Teng L, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A 2012; 109: 21360-5. ArticlePubMedPMC
REFERENCES
Figure & Data
References
Citations

- HOTAIR in cancer: diagnostic, prognostic, and therapeutic perspectives
Majid Nazari, Emad Babakhanzadeh, Arghavan Mollazadeh, Mohadese Ahmadzade, Elham Mohammadi Soleimani, Elnaz Hajimaqsoudi
Cancer Cell International.2024;[Epub] CrossRef - Long noncoding RNAs (lncRNAs) in human lymphomas
Ali Gholami, Khosro Farhadi, Fatemeh Sayyadipour, Masoud Soleimani, Fakhredin Saba
Genes & Diseases.2022; 9(4): 900. CrossRef - Long noncoding RNAs (lncRNAs) in HIV-mediated carcinogenesis: Role in cell homeostasis, cell survival processes and drug resistance
Lilian Makgoo, Salerwe Mosebi, Zukile Mbita
Non-coding RNA Research.2022; 7(3): 184. CrossRef - Biomedical impact of the expression of HOX locus-associated LncRNAs HOTAIR and HOTTIP in diffuse large B cell lymphoma
Mona Salah Eldin Habieb, Suzy Fawzy Goher, Abd-Elmonem Abd-Elkader El-Torgman, Ibrahim El Tantawy El Sayed, Najlaa Zanati Ali Abd-Elfattah
Human Gene.2022; 34: 201112. CrossRef - Mechanism of LncHOTAIR Regulating Proliferation, Apoptosis, and Autophagy of Lymphoma Cells through hsa-miR-6511b-5p/ATG7 Axis
Fu Gui, Xinyi Yu, Yemeng Wu, Chao Wu, Yulan Zhang, Peng-Yue Zhang
Evidence-Based Complementary and Alternative Medicine.2022; 2022: 1. CrossRef -
Circulating RNA biomarkers in diffuse large B-cell lymphoma: a systematic review
Philippe Decruyenaere, Fritz Offner, Jo Vandesompele
Experimental Hematology & Oncology.2021;[Epub] CrossRef - Circulating long non-coding RNAs HOTAIR, Linc-p21, GAS5 and XIST expression profiles in diffuse large B-cell lymphoma: association with R-CHOP responsiveness
Mahmoud A. Senousy, Aya M. El-Abd, Raafat R. Abdel-Malek, Sherine M. Rizk
Scientific Reports.2021;[Epub] CrossRef - An immunotherapeutic approach to decipher the role of long non-coding RNAs in cancer progression, resistance and epigenetic regulation of immune cells
Krishnapriya M. Varier, Hemavathi Dhandapani, Wuling Liu, Jialei Song, Chunlin Wang, Anling Hu, Yaacov Ben-David, Xiangchun Shen, Yanmei Li, Babu Gajendran
Journal of Experimental & Clinical Cancer Research.2021;[Epub] CrossRef - Cancer‑associated fibroblast‑derived CCL5 contributes to cisplatin resistance in A549 NSCLC cells partially through upregulation of lncRNA HOTAIR expression
Xiangjun Sun, Zhijie Chen
Oncology Letters.2021;[Epub] CrossRef - Competitive Endogenous RNA Network Involving miRNA and lncRNA in Non-Hodgkin Lymphoma: Current Advances and Clinical Perspectives
Mara Fernandes, Herlander Marques, Ana Luísa Teixeira, Rui Medeiros
Biomedicines.2021; 9(12): 1934. CrossRef - EZH2 expression is dependent on MYC and TP53 regulation in diffuse large B‐cell lymphoma
Eduardo Henrique Neves Filho, Carlos Gustavo Hirth, Igor Allen Frederico, Rommel Mario Burbano, Thiago Carneiro, Silvia Helena Rabenhorst
APMIS.2020; 128(4): 308. CrossRef Long Noncoding RNAs in Diffuse Large B-Cell Lymphoma: Current Advances and Perspectives
Xianbo Huang, Wenbin Qian, Xiujin Ye
OncoTargets and Therapy.2020; Volume 13: 4295. CrossRef- Lnc SMAD5-AS1 as ceRNA inhibit proliferation of diffuse large B cell lymphoma via Wnt/β-catenin pathway by sponging miR-135b-5p to elevate expression of APC
Chen-Chen Zhao, Yang Jiao, Yi-Yin Zhang, Jie Ning, Yi-Ruo Zhang, Jing Xu, Wei Wei, Gu Kang-Sheng
Cell Death & Disease.2019;[Epub] CrossRef - H3K18Ac as a Marker of Cancer Progression and Potential Target of Anti-Cancer Therapy
Marta Hałasa, Anna Wawruszak, Alicja Przybyszewska, Anna Jaruga, Małgorzata Guz, Joanna Kałafut, Andrzej Stepulak, Marek Cybulski
Cells.2019; 8(5): 485. CrossRef - HOTAIR as a Prognostic Predictor for Diverse Human Cancers: A Meta- and Bioinformatics Analysis
Halil Ibrahim Toy, Didem Okmen, Panagiota I. Kontou, Alexandros G. Georgakilas, Athanasia Pavlopoulou
Cancers.2019; 11(6): 778. CrossRef - Long Noncoding RNA HOTAIR Promotes Endometrial Carcinoma Cell Proliferation by Binding to PTEN via the Activating Phosphatidylinositol 3-Kinase/Akt Signaling Pathway
Xiao-Hui Zhang, Pin Hu, Yang-Qin Xie, Yong-Jun Kang, Min Li
Molecular and Cellular Biology.2019;[Epub] CrossRef - EZH2 abnormalities in lymphoid malignancies: underlying mechanisms and therapeutic implications
Boheng Li, Wee-Joo Chng
Journal of Hematology & Oncology.2019;[Epub] CrossRef - The prognostic impact of long noncoding RNA HOTAIR in leukemia and lymphoma: a meta-analysis
Yun Lin, Zhihong Fang, Zhijuan Lin, Zhifeng Li, Jintao Zhao, Yiming Luo, Bing Xu
Hematology.2018; 23(9): 600. CrossRef - Retracted: Downregulation of Long Noncoding RNA HOTAIR and EZH2 Induces Apoptosis and Inhibits Proliferation, Invasion, and Migration of Human Breast Cancer Cells
Lu Han, Hai-Chao Zhang, Li Li, Cai-Xia Li, Xu Di, Xin Qu
Cancer Biotherapy and Radiopharmaceuticals.2018; 33(6): 241. CrossRef - Long Non-Coding RNAs Guide the Fine-Tuning of Gene Regulation in B-Cell Development and Malignancy
Mette Dahl, Lasse Sommer Kristensen, Kirsten Grønbæk
International Journal of Molecular Sciences.2018; 19(9): 2475. CrossRef - HOTAIR, a long noncoding RNA, is a marker of abnormal cell cycle regulation in lung cancer
Minghui Liu, Hongyi Zhang, Ying Li, Rui Wang, Yongwen Li, Hongbing Zhang, Dian Ren, Hongyu Liu, Chunsheng Kang, Jun Chen
Cancer Science.2018; 109(9): 2717. CrossRef - The evolving concept of cancer stem-like cells in thyroid cancer and other solid tumors
Heather Hardin, Ranran Zhang, Holly Helein, Darya Buehler, Zhenying Guo, Ricardo V Lloyd
Laboratory Investigation.2017; 97(10): 1142. CrossRef - Emerging roles for long noncoding RNAs in B-cell development and malignancy
M. Winkle, J.L. Kluiver, A. Diepstra, A. van den Berg
Critical Reviews in Oncology/Hematology.2017; 120: 77. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure



Fig. 1.
Fig. 2.
Fig. 3.
| Variable | No. (%) (n = 164) | HOTAIR |
p-value | |
|---|---|---|---|---|
| HOTAIRlow | HOTAIRhigh | |||
| Sex | ||||
| Male | 98 (59.8) | 73 (58.4) | 25 (64.1) | .526 |
| Female | 66 (40.2) | 52 (41.6) | 14 (35.9) | |
| Age (yr) | ||||
| ≤ 60 | 94 (57.3) | 69 (55.2) | 25 (64.1) | .326 |
| > 60 | 70 (42.7) | 56 (44.8) | 14 (35.9) | |
| ECOG performance status |
||||
| < 2 | 132 (80.5) | 96 (76.8) | 36 (92.3) | .033 |
| ≥ 2 | 32 (19.5) | 29 (23.2) | 3 (7.7) | |
| Ann-Arbor stage |
||||
| I–II | 89 (54.6) | 63 (50.8) | 26 (66.7) | .083 |
| III–IV | 74 (45.4) | 61 (49.2) | 13 (33.3) | |
| Extranodal sites |
||||
| < 2 | 108 (66.3) | 81 (65.3) | 27 (69.2) | .653 |
| ≥ 2 | 55 (33.7) | 43 (34.7) | 12 (30.8) | |
| Lactate dehydrogenase |
||||
| Normal | 90 (55.2) | 65 (52.4) | 25 (64.1) | .201 |
| Elevated | 73 (44.8) | 59 (47.6) | 14 (35.9) | |
| IPI risk group |
79 (63.7) | 28 (71.8) | .354 | |
| 0–2 | 107 (65.6) | 45 (36.3) | 11 (28.2) | |
| 3–5 | 56 (34.4) | |||
| Hans classification |
||||
| GCB | 44 (28.0) | 32 (27.1) | 12 (30.8) | .660 |
| Non-GCB | 113 (72.0) | 86 (72.9) | 27 (69.2) | |
| EBV | ||||
| Negative | 132 (88.0) | 98 (87.5) | 34 (89.5) | .746 |
| Positive | 18 (12.0) | 14 (12.5) | 4 (10.5) | |
| BCL2 |
||||
| Negative expression | 84 (58.7) | 58 (54.7) | 26 (70.3) | .098 |
| Positive expression | 59 (41.3) | 48 (45.3) | 11 (29.7) | |
| c-MYC |
||||
| Negative expression | 92 (62.2) | 63 (57.3) | 29 (76.3) | .037 |
| Positive expression | 56 (32.8) | 47 (42.7) | 9 (23.7) | |
| c-MYC/BCL2 |
||||
| No co-expression | 110 (77.5) | 76 (72.4) | 34 (91.9) | .015 |
| Co-expression | 32 (22.5) | 29 (27.6) | 3 (8.1) | |
| Variable | Category | Univariate analysis p-value | Multivariate analysis p-value | HR | 95% CI |
|---|---|---|---|---|---|
| Age | > 60 vs ≤ 60 | < .001 | - | - | - |
| ECOG PS | ≥ 2 vs < 2 | < .001 | - | - | - |
| Ann-Arbor stage | III–IV vs I–II | .003 | - | - | - |
| Extranodal sites | ≥ 2 vs < 2 | .001 | - | - | - |
| Lactate dehydrogenase | Elevated vs normal | < .001 | - | - | - |
| IPI risk group | 3–5 vs 0–2 | < .001 | < .001 | 3.5 | 1.9–6.3 |
| Hans classification | Non-GCB vs GCB | .16 | - | - | - |
| EBV | Positive vs negative | .611 | - | - | - |
| BCL2 | Positive vs negative | .076 | - | - | - |
| c-MYC | Positive vs negative | .032 | - | - | - |
| c-MYC/BCL2 | Co-expression vs no co-expression | .006 | .528 | 1.2 | 0.6–2.4 |
| EZH2 | High vs non-high | .883 | - | - | - |
| SUZ12 | High vs non-high | .028 | .183 | 1.5 | 0.8–2.7 |
| EED | High vs non-high | .25 | - | - | - |
| H3K27me3 | High vs non-high | .01 | .023 | 2 | 1.1–3.6 |
| HOTAIR | High vs non-high | .004 | .086 | 0.5 | 0.2–1.1 |
Values are presented as number (%). HOTAIR, hox transcript antisense intergenic RNA; DLBCL, diffuse large B-cell lymphoma; ECOG, Eastern Cooperative Oncology Group; IPI, International Prognostic Index; GCB, germinal center B-like; EBV, Epstein-Barr virus. Information not available in some cases.
HR, hazard ratio; CI, confidence interval; ECOG PS, Eastern Cooperative Oncology Group performance status; IPI, International Prognostic Index; EBV, Epstein-Barr virus; EZH2, enhancer of zeste homolog 2; SUZ12, suppressor of zeste 12 homolog; EED, embryonic ectoderm development; HOTAIR, hox transcript antisense intergenic RNA.

 E-submission
E-submission