Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 51(3); 2017 > Article
-
Review
Molecular Testing of Lung Cancers - Hyo Sup Shim, Yoon-La Choi1, Lucia Kim2, Sunhee Chang3, Wan-Seop Kim4, Mee Sook Roh5, Tae-Jung Kim6, Seung Yeon Ha7, Jin-Haeng Chung8, Se Jin Jang9, Geon Kook Lee,10, , The Korean Cardiopulmonary Pathology Study Group
-
Journal of Pathology and Translational Medicine 2017;51(3):242-254.
DOI: https://doi.org/10.4132/jptm.2017.04.10
Published online: April 21, 2017
Department of Pathology, Yonsei University College of Medicine, Seoul, Korea
1Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
2Department of Pathology, Inha University School of Medicine, Incheon, Korea
3Department of Pathology, Inje University Ilsan Paik Hospital, Inje University, Goyang, Korea
4Department of Pathology, Konkuk University School of Medicine, Seoul, Korea
5Department of Pathology, Dong-A University College of Medicine, Busan, Korea
6Department of Pathology, College of Medicine, The Catholic University of Korea, Seoul, Korea
7Department of Pathology, Gachon University Gil Medical Center, Incheon, Korea
8Department of Pathology, Seoul National University Bundang Hospital, Seongnam, Korea
9Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
10Department of Pathology, National Cancer Center, Goyang, Korea
- Corresponding Author Geon Kook Lee, MD Department of Pathology, National Cancer Center, 323 Ilsan-ro, Ilsandong-gu, Goyang 10408, Korea Tel: +82-31-920-1746 Fax: +82-31-920-1369 E-mail: gklee@ncc.re.kr
© 2017 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
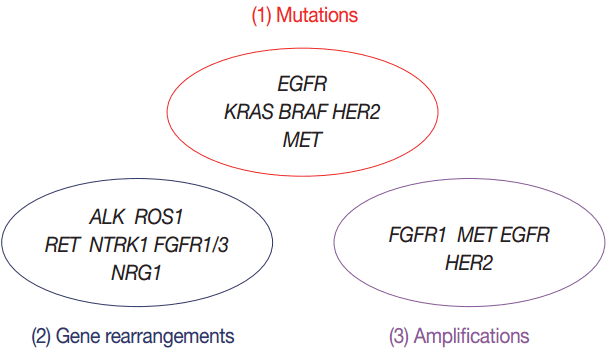
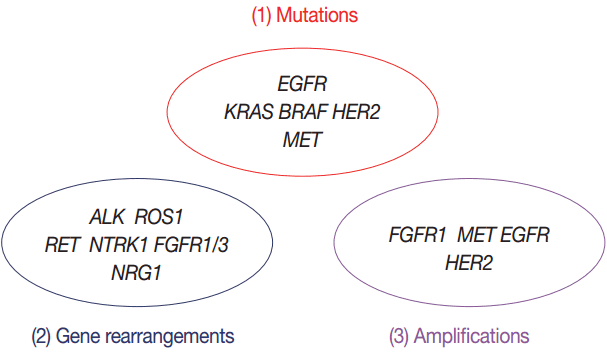
- Targeted therapies guided by molecular diagnostics have become a standard treatment of lung cancer. Epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK) rearrangements are currently used as the best predictive biomarkers for EGFR tyrosine kinase inhibitors and ALK inhibitors, respectively. Besides EGFR and ALK, the list of druggable genetic alterations has been growing, including ROS1 rearrangements, RET rearrangements, and MET alterations. In this situation, pathologists should carefully manage clinical samples for molecular testing and should do their best to quickly and accurately identify patients who will benefit from precision therapeutics. Here, we grouped molecular biomarkers of lung cancers into three categories—mutations, gene rearrangements, and amplifications—and propose expanded guidelines on molecular testing of lung cancers.
- Mutations including EGFR, KRAS, BRAF, HER2, and MET
- Activating somatic mutations including point substitution, small insertion, and in-frame deletion are major oncogenic drivers in lung cancer. The discovery of activating EGFR mutations and their close relation with the response to EGFR TKIs opened the new era of precision medicine. Since then, clinical trials have confirmed that EGFR mutations are the best predictive factor of EGFR TKI efficacy [7,8]. EGFR mutations have been more frequently found in lung adenocarcinomas in Asian populations than Western populations [9]. The overall EGFR mutation rate is approximately up to 46% among Korean patients with adenocarcinoma [10,11]. More than 90% of sensitizing EGFR mutations are composed of an in-frame deletion in exon 19 and a point mutation (L858R) in exon 21. Nonetheless, most patients experience disease progression usually after 12 months of treatment [12,13]. A variety of mechanisms are involved in acquired resistance to EGFR TKIs [12-14]. Among these, the most common resistance mechanism is the T790M mutation in EGFR, constituting 50%–60% of cases [12,13,15]. Recently, osimertinib was approved for the treatment of T790M-positive lung cancers in Korea [16]. The acquired EGFR C797S mutation has been reported to mediate resistance to osimertinib in a subset of EGFR T790M–positive non-small cell lung carcinomas (NSCLCs) [17,18]. Thus, EGFR mutation testing covering sensitizing as well as nonsensitizing mutations has become the most important step in treatment decision-making for lung cancer patients because of the high frequency of EGFR mutations and the availability of targeted therapeutic agents.
- The frequency of KRAS mutations has been reported to be 8%–15% in lung adenocarcinoma among Korean patients [19,20]. Unfortunately, targeted therapies thus far have been unsuccessful [21]. KRAS mutations are known as negative predictive markers of a poor response to EGFR TKIs. In addition, a KRAS mutation has been reported to be a poor prognostic factor. This also appears to be dependent both on the specific KRAS codon mutation and disease stage at the time of diagnosis [21]. Invasive mucinous adenocarcinomas show a high incidence of KRAS mutations [22].
- Besides EGFR mutations, other targetable mutations have been found although their frequency is low. A BRAF mutation has been found in 3%–4% of lung adenocarcinomas in Western patients [23,24]. Among never-smoker Korean women with lung cancer, the frequency of tumor BRAF mutation is 1% [25]. The V600E mutation constitutes only 50% of BRAF mutations in contrast to other cancers (such as melanoma and papillary thyroid carcinoma) [23,24]. Vemurafenib and dabrafenib show clinical activity in BRAF V600E-mutant NSCLC [26-28]. HER2 mutations are present in approximately 1%–2% of lung adenocarcinomas [29]. The most common mutation is an in-frame insertion within exon 20 [29]. The exon 20 insertion results in increased HER2 kinase activity. The clinical response to HER2-targeting agents, such as trastuzumab and afatinib, is observed in patients with lung cancer harboring a HER2 mutation [29].
- Recently, MET splice site mutations have emerged as targetable oncogenic drivers. Oncogenic mutations in the MET exon 14 splice sites (these mutations cause exon 14 skipping) occur in 3%–4% of lung adenocarcinomas [30,31]. Patients with lung cancer harboring MET exon 14 skipping show a clinical response to MET inhibitors including crizotinib [30,31].
- The important reason for molecular testing of lung cancers is to select patients who may benefit from targeted therapies. In addition, patients with lung cancer can get benefits from molecular testing of their tumors regardless of stage. For example, molecular testing can provide accurate information on staging (in case of multiple tumors), prognostic stratification, and prompt treatment in case of recurrence. Because of the high frequency of EGFR mutations in Asian populations as mentioned above, EGFR mutation testing is especially important for the treatment of Korean lung cancer patients. The Korean Food and Drug Administration (FDA) approved only EGFR molecular testing from among five genes important for decision-making with respect to first-line chemotherapy during standard treatment. Other mutations can also be approved and used as predictive markers in the near future. Each mutation is significantly associated with some clinical factors or histological subtypes. Nevertheless, clinical findings alone cannot completely predict specific mutation status [32,33]. In most of the guidelines published so far, histological types have been recommended as the most important factor in determining whether to perform molecular tests [34-36]. In particular, for patients who have a diagnosis of NSCLC with an adenocarcinoma component or nonsquamous cell type, molecular testing is routinely recommended [35]. Thus, pathologists should try to further classify NSCLCs into more specific subtypes, such as adenocarcinoma or squamous cell carcinoma (SQC) [2]. In addition, a minimum immunohistochemical panel, such as one adenocarcinoma marker and one SQC marker, is recommended to preserve as much tissue as possible for molecular testing in small tissue samples [35,37]. Besides histological analysis, in cases of young age, female gender, never-smokers, small biopsies, or patients with a combined tumor type, molecular testing can be done.
- A variety of methods can be used for detecting mutations including direct sequencing, real-time polymerase chain reaction (PCR), and commercial kits (Table 2) [38,39]. Pathologists should consider the available approaches and the advantages and disadvantages of each method, including analytical sensitivity and turnaround time. In addition, new technologies for molecular testing must be approved by the Korean government before incorporation into clinical practice.
- Direct sequencing is considered the gold standard for mutation analysis. On the other hand, direct DNA sequencing requires a high ratio of tumor tissue to normal tissue (more than 50% tumor content) for reliable results. In contrast, PCR-based methods show high sensitivity, requiring mutant DNA content of only 1% [38]. Nevertheless, these PCR-based methods can detect only previously known mutations or targeted sites. PCR-based methods including the peptide nucleic acid–mediated PCR clamping method and allele-specific PCR method have been approved in Korea. Among the targetable mutations, EGFR mutations have been the most studied in the field of lung cancer. In studies comparing two representative methods, clinical outcomes are not significantly different among groups harboring EGFR mutations detected by direct sequencing or by PCR-based methods [40,41]. Highly sensitive methods may also be useful for the detection of EGFR mutations associated with acquired resistance, e.g., T790M [15]. As with the EGFR mutation, direct sequencing or real-time PCR can be used for the detection of KRAS, BRAF, and HER2 mutations, which include a point mutation or small indel. In the case of MET exon 14 alterations, most alterations are a point mutation or indel, affecting the splice acceptor site, splice donor site, or the approximately 25-base pair (bp) intronic noncoding region. In rare cases, however, whole-exon deletions of MET exon 14 have been reported [42]. A DNA-based method including direct sequencing is possible, but the mRNA-based method including real-time PCR is better at detecting MET exon 14 skipping.
- As targetable genetic alterations are increasingly discovered, individual genotyping may become relatively inefficient and costly. Thus, next-generation sequencing (NGS) technology with DNA or RNA is reported to be useful for multiplexed and deep genomic sequencing [43,44]. Especially, targeted deep sequencing of selected gene sets (a so-called cancer panel) is approaching integration into daily clinical practice. Nonetheless, due to the methodological complexity of NGS, analytical validation and quality control are crucial for implementation in routine practice. Furthermore, to become a companion diagnostic assay, an NGS-based method should be comparatively validated with previous companion diagnostics.
- As previously described in our proposed guideline, small biopsy and cytology samples are used for mutation testing [2]. In particular, all samples obtained from a variety of methods are acceptable, including transbronchial lung biopsy, endobronchial ultrasound-guided transbronchial needle aspiration, bronchial brushing or washing, computed tomography–guided gun biopsy or needle aspiration, and pleural fluid sampling [34,45,46]. The results using cytology samples are highly concordant with those of the corresponding tissue samples, especially with more sensitive methods [45,47].
- Since most driver mutations develop in early steps of carcinogenesis, tumor samples from either primary masses or metastatic lesions are equally suitable for mutation testing [48]. In the case of multiple primary lung cancers, each tumor may be tested [49].
- Formalin-fixed, paraffin-embedded (FFPE) tissues are most frequently used in molecular testing. Routinely prepared FFPE tissues are the most practical resource for molecular analysis, in spite of fixation-related artifacts [50]. The optimal fixative for preparing FFPE samples is generally 10% neutral-buffered formalin [34]. The optimal fixation time ranges from 6 to 72 hours to avoid underfixation or overfixation, respectively [34,50]. Routinely prepared cytology samples such as alcohol-fixed smears or Thin Prep slides [45,47] and cell block samples [51] are also suitable for mutation testing.
- Because obtaining a tissue sample from bronchoscopy or a transthoracic needle biopsy is difficult and nonrepeatable, a liquid biopsy, which contains circulating tumor DNA, circulating tumor cells, or exosomes using plasma or body fluids, has recently been attracting attention as a new source for identification of somatic mutations [52,53]. Nonetheless, due to the high rate of false negative results at present, negative results require a tissue biopsy to determine exact mutation status [53].
- Pathologists must confirm the presence of tumor cells in a sample before mutation analysis. The percentage and quality of tumor cells are crucial for proper mutation testing. For example, direct sequencing requires at least 20% tumor cells in the sample for reliable testing result. Thus, evaluation of the percentage of tumor cells (tumor purity) in a given sample is highly recommended, especially for less sensitive methods. Tumor dissection may be used to increase the tumor content if required. Thus, as noted, the minimum number or percentage of cancer cells required for adequate testing depends on the analytic sensitivity of the testing method.
- Mutation testing reports should contain identification of the patient (pathology number, hospital unit number, age, and gender), requesting physician with department, receipt day, report day, sample used for the testing (site or organ, sample type, and tumor purity), methodology used, exons tested, test results, comments on results, and names of the testing technician and corresponding pathologist [2].
- Analytical and clinical validation should be done when the testing is implemented in the laboratory. One of the references is indexes of evaluation from the Korean Institute of Genomic Testing Evaluation.
- Repeat examination can be considered in cases of poor sequencing data, results not matching previously well-defined clinical features or a unique histological subtype, or concurrent detection of mutually exclusive driver mutations. Other types of methods may be useful in equivocal cases.
- For optimal mutation testing, the quality of a sample and validation status of the testing method are crucial. Laboratories must incorporate mutation testing methods into their overall laboratory quality improvement program, by establishing appropriate quality improvement monitors as needed to ensure consistent performance at all steps of the testing and reporting processes. In particular, laboratories should participate in a formal proficiency testing program, if available, or an alternative proficiency assurance activity. There is an external quality control program for mutation testing from the Korean Institute of Genomic Testing Evaluation and the Korean Society of Pathologists. To improve the reliability of assays in terms of detection of mutations, a regular quality control program should be implemented, such as examination of reference materials or comparison of the results between different diagnostic assays.
- Gene rearrangements including ALK, ROS1, RET, NTRK1, NRG1, and FGFR
- Since the discovery of a transforming fusion gene between echinoderm microtubule-associated protein-like 4 (EML4) and ALK in 2007, the list of targetable gene fusions in lung cancer has been growing [54,55]. Although the prevalence of each gene fusion is low (approximately 1%–5%), molecularly tailored treatments have resulted in dramatic clinical responses. The presence of an ALK rearrangement and ROS1 proto-oncogene receptor tyrosine kinase (ROS1) rearrangement in lung cancer has become the best predictor of response to crizotinib [56,57]. Subsequently, RET proto-oncogene (RET) fusions were found comprising 1% of lung adenocarcinomas [58,59]. Cabozantinib or vandetanib showed clinical activity against lung cancer harboring RET fusions [60,61]. Besides ALK, ROS1, and RET fusions, NTRK1 and fibroblast growth factor receptor (FGFR) 1/3 fusions were reported in lung cancer, which are expected to give a therapeutic opportunity to patients with no known driver alterations [62,63]. Of note, NRG1 fusions were predominantly found in invasive mucinous adenocarcinoma, which is a unique variant of lung adenocarcinoma [22,64].
- Clinical characteristics associated with gene fusions such as ALK, ROS1, RET, and NTRK1 are adenocarcinoma histological features, never- or light-smoking history, and younger age. Nevertheless, not all gene fusions have these characteristics. FGFR fusions are associated with smoking history [63]. Therefore, clinical characteristics alone cannot determine gene rearrangement testing.
- Histological type is an important factor in determining whether to perform genetic analysis or which genes to test [36]. Most targetable gene fusions are discovered in NSCLCs. Because ALK, ROS1, and RET fusions are predominantly found in adenocarcinomas or NSCLCs with an adenocarcinoma component, pathologists should try to further classify NSCLCs into more specific subtypes. On the other hand, FGFR fusions can be found in adenocarcinomas and SQCs [63,65].
- Several methods are currently available for assessing gene rearrangement, including fluorescence in situ hybridization (FISH), immunohistochemistry (IHC), reverse-transcriptase polymerase chain reaction (RT-PCR), and NGS (Table 2).
- Break apart FISH is the currently standard method for the detection of gene rearrangements. Although the FISH assay requires technical expertise and experience for interpretation, this assay is reasonably sensitive and specific for the detection of gene rearrangements regardless of fusion partners. The pathologist should recognize the probe design, especially which sides are labeled with a red and green signal and the positive criteria. Crizotinib, a first generation ALK inhibitor, was approved by the United States FDA in conjunction with a companion diagnostic FISH test (ALK Break Apart FISH Probe Kit, Abbott Molecular Inc.). Positive criteria for the Abbott FISH probe kit are as follows. Positive signals are defined as separate signals or a single red signal. Initially, 50 tumor cells are counted. A sample is considered positive if > 25 cells out of 50 (> 25/50 or > 50%) test positive. A sample is considered equivocal if 5 to 25 cells (10 to 50%) test positive. If the sample is equivocal, a second reader should evaluate the slide. The first and second cell count readings are added up, and a percentage is calculated out of 100 cells. If the percentage of positive cells is ≥ 15% (≥ 15/100), the sample is considered positive. The Korean FDA also approved the ALK FISH as a selection test for crizotinib treatment in lung cancer patients.
- Because protein expression can serve as a surrogate marker of gene rearrangements, IHC is useful for screening of gene rearrangements when it is appropriately validated [3,66-68]. In particular, ALK IHC was extensively studied and validated [3]. Among ALK IHC antibodies, 5A4 and D5F3 appear to be superior to the ALK1 antibody for detecting ALK-rearranged lung cancer [69,70]. IHC can effectively predict ALK fusion status because lung adenocarcinoma in general does not express the ALK protein without genetic alterations. The VENTANA ALK (D5F3) CDx Assay was approved by the U.S. FDA for identification of patients eligible for treatment with crizotinib. Other antibodies are not validated as companion diagnostics; therefore, their use should be limited to a screening step within controlled settings.
- Besides ALK, there are no approved companion diagnostics for other gene rearrangements. Break apart FISH is generally used to detect gene arrangements in clinical samples, and IHC can serve as a screening method.
- In terms of mRNA expression, the RT-PCR assay is used to find fusion transcripts with unique sequences. RT-PCR methods are generally known to be sensitive [71]. Nonetheless, RT-PCR has some limitations in clinical practice. Primer design requires information about fusion partners and breakpoints. Thus, only known fusion variants can be detected. In addition, most materials for molecular testing are FFPE samples, where RNAs can be severely degraded compared to fresh frozen tissue. Nevertheless, RT-PCR can be useful when only cytology samples are available [71].
- The NGS technology has emerged as an alternative to the detection of gene rearrangements. It can simultaneously detect a variety of gene fusions using targeted DNA sequencing or RNA sequencing [43,44]. The sensitivity and specificity for calling a gene fusion mainly depend on the detection algorithm. The NGS-based gene rearrangement method should be vigorously validated before implementation in routine practice. Furthermore, to become a companion diagnostic test, an NGS-based method should be comparatively validated with previous companion diagnostics.
- A variety of biopsy specimens can be used for gene rearrangement testing as described above in the section on mutation testing. Previous studies have shown that cytology samples are suitable for gene rearrangement analysis, especially as validated in ALK fusion testing [72]. The cell block is usually first used if there are enough tumor cells in the block. Smear slides can also be used for the detection of gene rearrangement [72]. Thus, both tissue and cytology samples are acceptable for gene rearrangement testing, if appropriately validated. Details were described in the previous guideline [3].
- For adequate evaluation of break apart FISH, a minimum of 50 to 100 well-preserved tumor cells are required. Tumor purity is not as critical for FISH tests, however, it is necessary to choose areas of a slide in which the viable tumor cells are most abundant. Because a dark-field fluorescence microscope does not provide perfect visualization of tumor morphology, the pathologist should determine whether the selected area contains a sufficient number of tumor cells and should interppret the result carefully by distinguishing between cancer cells and intimately mixed non-neoplastic cells.
- As for RT-PCR and NGS technologies, the tumor percentage can be critical in gene rearrangement testing as described in mutation testing. The pathologist should verify the quality of the sample and the presence of tumor cells. Tumor dissection may be used to increase the tumor content if required.
- Reports on gene rearrangement testing should contain identification of the patient (pathology number, hospital unit number, age, and gender), requesting physician with department, receipt day, report day, sample used for the testing (site or organ, sample type, and tumor purity), methodology used, detectable fusion variants, test results (negativity or positivity for the gene rearrangement), comments on results, and names of the testing technician and corresponding pathologist. We recommend reporting the additional details in the case of a FISH test: information on probe design, total number of counted nuclei, and percentage of the tumor nuclei showing positive signals (split signal or isolated signal containing a kinase domain) [3].
- General principles are similar to those of mutation testing. Technical and clinical validation procedures should be carried out when the testing is set up in the laboratory. One of the references is indexes of the evaluation from the Korean Institute of Genomic Testing Evaluation.
- Pathologists should recognize the limitations of each methodology. With respect to the break apart FISH test, repeat examination can be considered in cases of poor signals, atypical signal patterns (including predominantly isolated 5' signals), a borderline range that approached the 15% cutoff line, results not matching well-validated IHC, or concurrent detection of driver mutations. Other types of methods may be useful in equivocal cases.
- There is an external quality control program for ALK fusion testing from the Korean Institute of Genomic Testing Evaluation and the Korean Society of Pathologists. To improve the reliability of assays for detecting gene rearrangements, a regular quality control program should be performed, and the results of molecular testing can be confirmed by different diagnostic assays, such as validated IHC.
- Amplifications including FGFR1, EGFR, MET, and HER2
- Gene amplification is an important mechanism for oncogene activation. HER2-targeting therapy has been implemented in breast and gastric cancer on the basis of molecular testing for HER2. Amplifications of several genes have been studied in lung cancer.
- FGFR1 is one of the most commonly amplified genes in human cancers. Amplified FGFR1 has been described in 13%–22% of lung SQCs [73,74]. It is expected to become a promising targetable alteration in lung SQCs because there are no known targeted therapeutic agents for SQCs [73]. In vivo studies have revealed that inhibition of the FGFR1 pathway with FGFR inhibitors leads to significant tumor shrinkage, suggesting that FGFR inhibitors might be an effective therapeutic option in SQCs with FGFR1 amplification [75]. Nevertheless, clinical trials of FGFR inhibitors were not impressive [76]. Additional biomarkers should be studied to predict the drug response to FGFR inhibitors [77]. A study showed that homogeneous high-level amplification was associated more strongly with a clinical response to FGFR inhibition [78]. Thus, FGFR1 amplification can still be a promising candidate for targeted therapy.
- EGFR amplifications were found in adenocarcinomas and SQCs. Initially, EGFR amplification was regarded as a predictive biomarker of EGFR TKI treatment [79]. Subsequent studies revealed that EGFR amplification was closely related to EGFR mutation, and EGFR mutation was the strongest predictive marker of the response to EGFR TKIs [7,80]. Among SQCs, EGFR high copy number gains/amplifications were present in 8%–9% of cases [4,81]. Patients with lung SQC harboring an EGFR high copy number gains or amplifications showed a higher response rate to EGFR TKI [81].
- De novo MET amplification was present in 2%–4% of lung adenocarcinomas [5,82]. NSCLC patients with de novo MET amplification showed a rapid and durable response to crizotinib [83,84]. MET amplification was also identified as one of the acquired secondary resistance mechanisms in patients with EGFR mutations who progressed on EGFR TKIs [12,13].
- De novo HER2 amplification was found to be present in 1%–2% of lung adenocarcinomas [5]. NSCLC patients with de novo HER2 amplification can be treated with HER2-targeting agents [85]. HER2 amplification was also identified as one of the acquired secondary resistance mechanisms in patients with EGFR mutations who progress on EGFR TKIs [18,86].
- Amplifications of receptor tyrosine kinase genes are present in lung cancers regardless of histological subtypes, while the frequencies are different. Thus, these gene amplifications can be examined in patients with lung cancer, especially previously targetable oncogene-negative cases. FGFR1 amplification is especially meaningful for SQCs.
- EGFR amplification can serve as a predictive marker of SQCs. MET and HER2 amplification can be examined in the primary tumors or tumors with acquired resistance to TKIs.
- Several methods can be used for detection of gene amplifications (Table 2). FISH using a locus-specific intensifier (LSI) gene and a chromosome-specific centromere (CEP) probe is a standard method for the detection of gene amplifications. Although the FISH assay requires technical expertise and experience for interpretation, this method has the advantage of being able to evaluate gene amplification by selecting only cancer cells.
- FISH-positive criteria have been used according to the corresponding genes. FGFR1 high-level amplification is defined as copy number ≥ 9 or (1) an FGFR1/CEP8 ratio of ≥ 2.0, (2) the average number of FGFR1 signals per tumor cell nucleus ≥ 6, and (3) the percentage of tumor cells containing ≥ 15 FGFR1 signals or large clusters ≥ 10% [74,87]. According to previously published criteria, the EGFR gene copy number was classified into six FISH strata: disomy (two or fewer copies in more than 90% of cells), low trisomy (two or fewer copies in 40% or more of the cells, three copies in 10%–40% of cells, and four or more copies in less than 10% of cells), high trisomy (two or fewer copies in ≥ 40% of cells, three copies in ≥ 40% of cells, and four or more copies in less than 10% of cells), low polysomy (four or more copies in 10%–40% of cells), high polysomy (four or more copies in 40% of the cells or more), and gene amplification (defined by the presence of tight EGFR clusters and a ratio of EGFR to the chromosome of ≥ 2.0, or ≥ 15 copies of EGFR per cell in ≥ 10% of the cells analyzed) [79]. Regarding MET, FISH-positive groups include (1) high-level amplification (presence of loose or tight clusters of MET signals too numerous to count) or a MET/CEP7 ratio greater than 5.0 and (2) low-level amplification (tumors with the MET/CEP7 ratio ≥ 2.2 and ≤ 5.0) [83]. As for HER2, gene amplification can be defined as positive when the HER2/CEP17 ratio is ≥ 2.0 or average HER2 copy number is ≥ 6.0 signals/cell based on the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) guidelines for dual-probe in situ hybridization [88].
- In clinical practice, for many years FISH has been the most widely used method for gene copy number assessment. Silver in situ hybridization (SISH) is a new technology with some clinical advantages over FISH. Examination can be made using conventional light microscopy with preserved tissue architecture. In addition, the slides are durable and can be reviewed several years after staining. Thus, the SISH technique can be more practically applied in routine diagnostic procedures [89].
- Quantitative real-time PCR can be used to detect gene amplification [90]. In this case, the optimal cutoff threshold of amplification should be determined considering clinical relevance and a response to targeted therapeutic agents.
- The NGS technology can simultaneously detect a variety of genes with a copy number gain using DNA sequencing [43]. Nonetheless, currently, the NGS algorithm for determination of a copy number gain in heterogeneous tumor tissue is not well established. The established guideline is based on a specific gene copy number, but the NGS technology cannot determine the specific gene number in given cancer cells. The reliability of testing results also depends on the tumor cell percentage and the degree of gene amplification. Before clinical implementation, thorough verification is required.
- Various biopsy samples can be used for amplification testing as in other molecular testing methods described above. Samples routinely consist of FFPE tissues. Tumor tissues from either primary tumors or metastatic lesions can be equally available for molecular testing especially regarding high-level homogeneous amplification [91]. Cytology samples can be used with quantitative real-time PCR or NGS.
- Regarding the in situ hybridization method, a minimum of 50 to 100 tumor cells are required. General principles are similar to those described above in the section on break apart FISH testing.
- Reports on gene amplification testing should contain identification of the patient (pathology number, hospital unit number, age, and gender), requesting physician with department, receipt day, report day, sample used for the testing (site or organ, sample type, and tumor purity), methodology used, test results (negativity or positivity for the gene amplification according to proposed criteria), comments on results, and names of the testing technician and corresponding pathologist. Additional information can be included in the case of in situ hybridization: the total number of counted nuclei, gene/CEP ratio, average copy number, or findings according to special diagnostic criteria.
- Analytical and clinical validation procedures should be conducted when the testing is set up in the laboratory. One of the references is indexes of evaluation from the Korean Institute of Genomic Testing Evaluation. General principles are similar to the ASCO/CAP guidelines for dual-probe in situ hybridization [88].
- There is an external quality control program for FISH/SISH testing (for example, HER2 amplification) from the Korean Institute of Genomic Testing Evaluation and the Korean Society of Pathologists. To improve the reliability of assays for detection of gene amplification, a regular quality control program should be implemented, and the results of molecular testing can be confirmed by different diagnostic assays, such as validated IHC for protein overexpression.
MOLECULAR TESTING
Background
Indications
Methodology
Methods
Sample types
Sample requirements
Reporting
Validation of a test
Quality assurance
Background
Indications
Methodology
Methods
Sample type
Sample requirements
Reporting
Validation of the test
Quality assurance
Background
Indications
Methodology
Method
Sample type
Sample requirements
Reporting
Validation of a test
Quality assurance
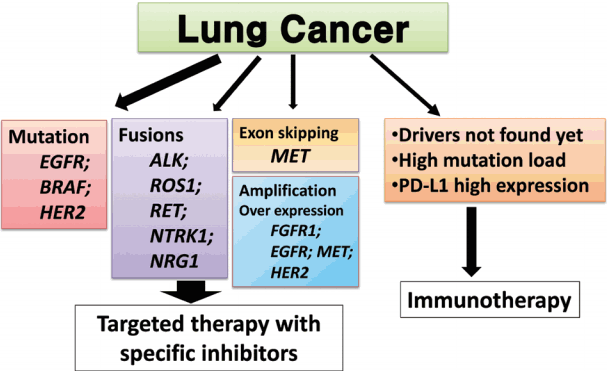
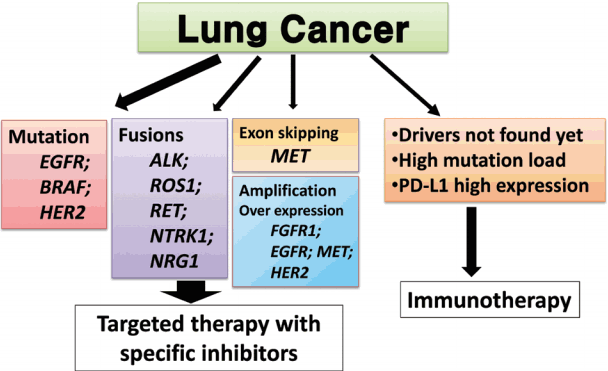
- We proposed guideline recommendations for molecular testing according to three categories of genetic alterations in lung cancer (summarized in Table 3). Molecular diagnostics for targetable genetic alterations have become the standard of care in the management of lung cancer patients (Fig. 2). In addition, immunotherapy has emerged as a new therapeutic option for patients who do not have targetable alterations (Fig. 2). Both therapeutic approaches require pathological diagnosis based on molecular biomarker assessment. Therefore, pathologists should carefully manage clinical samples for molecular testing and should do their best to quickly and accurately identify patients who will benefit from precision therapeutics. Each pathology department should fully validate the detection methods and should also participate in a quality control and formal proficiency testing program.
CONCLUSION
Acknowledgments
| Gene | Representative subtypes or variants | Frequency | Targeted agents |
|---|---|---|---|
| Mutations | |||
| EGFR | Exon 19 deletion, Exon 21 L858R, Exon 20 T790M | 40%–50% in ADCsa | Gefitinib, erlotinib, afatinib, osimertinib |
| 10%–20% in ADCsb | |||
| KRAS | G12X, G13X, G61X | 5%–10% in ADCsa | MEK inhibitors |
| 20%–30% in ADCsb | |||
| BRAF | V600E | 1%–4% in ADCs | Vemurafenib, dabrafenib, |
| HER2 | p.A775 G776insYVMA in exon 20 | 1%–2% in ADCs | Trastuzumab, afatinib |
| MET | Splice site mutations around or in exon 14 | 3%–4% in ADCs | Crizotinib, cabozantinib |
| Gene fusions | |||
| ALK | EML4-ALK, TGF-ALK, KIF5B-ALK | 5% in ADCs | Crizotinib, ceritinib, alectinib |
| ROS1 | CD74-ROS1, EZR-ROS1, SLC34A2-ROS1, SDC4-ROS1 | 1% in ADCs | Crizotinib, ceritinib |
| RET | KIF5B-RET, CCDC6-RET | 1% in ADCs | Cabozantinib, vandetanib, alectinib |
| NTRK1 | MPRIP-NTRK1 and CD74-NTRK1, TPM3-NTRK1 | < 1% in ADCs | Entrectinib |
| FGFR1/3 | FGFR3-TACC3, BAG4-FGFR1 | 1% in NSCLCs | FGFR inhibitor |
| NRG1 | CD74-NRG1, SLC3A2-NRG1, VAMP2-NRG1 | 7% in mucinous ADCs | NA |
| Amplifications | |||
| FGFR1 | Gene amplification | 13%–22% in SQCs | FGFR inhibitor |
| EGFR | Gene amplification | 8%–9% in SQCs, | EGFR inhibitor |
| MET | Gene amplification | 2%–4% in ADCs | Crizotinib |
| HER2 | Gene amplification | 1%–2% in ADCs | Trastuzumab, afatinib |
- 1. Li T, Kung HJ, Mack PC, Gandara DR. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol 2013; 31: 1039-49. ArticlePubMedPMC
- 2. Shim HS, Chung JH, Kim L, et al. Guideline recommendations for EGFR mutation testing in lung cancer: proposal of the Korean Cardiopulmonary Pathology Study Group. Korean J Pathol 2013; 47: 100-6. ArticlePubMedPMC
- 3. Kim H, Shim HS, Kim L, et al. Guideline recommendations for testing of ALK gene rearrangement in lung cancer: a proposal of the Korean Cardiopulmonary Pathology Study Group. Korean J Pathol 2014; 48: 1-9. ArticlePubMedPMC
- 4. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489: 519-25. ArticlePubMedPMCPDF
- 5. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014; 511: 543-50. ArticlePubMedPMCPDF
- 6. George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015; 524: 47-53. PubMedPMC
- 7. Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009; 361: 947-57. ArticlePubMed
- 8. Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010; 362: 2380-8. ArticlePubMed
- 9. Yatabe Y, Kerr KM, Utomo A, et al. EGFR mutation testing practices within the Asia Pacific region: results of a multicenter diagnostic survey. J Thorac Oncol 2015; 10: 438-45. ArticlePubMedPMC
- 10. Lee SH, Kim WS, Choi YD, et al. Analysis of mutations in epidermal growth factor receptor gene in Korean patients with non-small vell lung cancer: summary of a nationwide survey. J Pathol Transl Med 2015; 49: 481-8. ArticlePubMedPMCPDF
- 11. Lee B, Lee T, Lee SH, Choi YL, Han J. Clinicopathologic characteristics of EGFR, KRAS, and ALK alterations in 6,595 lung cancers. Oncotarget 2016; 7: 23874-84. ArticlePubMedPMC
- 12. Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011; 3: 75ra26.PubMedPMC
- 13. Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013; 19: 2240-7. ArticlePubMedPMCPDF
- 14. Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol 2014; 11: 473-81. ArticlePubMedPDF
- 15. Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res 2011; 17: 1169-80. ArticlePubMedPMCPDF
- 16. Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015; 372: 1689-99. ArticlePubMed
- 17. Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015; 21: 560-2. ArticlePubMedPMCPDF
- 18. Planchard D, Loriot Y, André F, et al. EGFR-independent mechanisms of acquired resistance to AZD9291 in EGFR T790M-positive NSCLC patients. Ann Oncol 2015; 26: 2073-8. ArticlePubMed
- 19. Lee B, Lee B, Han G, Kwon MJ, Han J, Choi YL. KRAS mutation detection in non-small cell lung cancer using a peptide nucleic acid-mediated polymerase chain reaction clamping method and comparative validation with next-generation sequencing. Korean J Pathol 2014; 48: 100-7. ArticlePubMedPMC
- 20. Kim HR, Ahn JR, Lee JG, et al. The impact of cigarette smoking on the frequency of and qualitative differences in KRAS mutations in Korean patients with lung adenocarcinoma. Yonsei Med J 2013; 54: 865-74. ArticlePubMedPMC
- 21. Wood K, Hensing T, Malik R, Salgia R. Prognostic and predictive value in KRAS in non-small-cell lung cancer: a review. JAMA Oncol 2016; 2: 805-12. ArticlePubMed
- 22. Shim HS, Kenudson M, Zheng Z, et al. Unique genetic and survival characteristics of invasive mucinous adenocarcinoma of the lung. J Thorac Oncol 2015; 10: 1156-62. ArticlePubMed
- 23. Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol 2011; 29: 3574-9. ArticlePubMed
- 24. Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol 2011; 29: 2046-51. ArticlePubMedPMC
- 25. Ha SY, Choi SJ, Cho JH, et al. Lung cancer in never-smoker Asian females is driven by oncogenic mutations, most often involving EGFR. Oncotarget 2015; 6: 5465-74. ArticlePubMedPMC
- 26. Planchard D, Besse B, Groen HJ, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 2016; 17: 984-93. ArticlePubMedPMC
- 27. Planchard D, Kim TM, Mazieres J, et al. Dabrafenib in patients with BRAF(V600E)-positive advanced non-small-cell lung cancer: a single-arm, multicentre, open-label, phase 2 trial. Lancet Oncol 2016; 17: 642-50. ArticlePubMedPMC
- 28. Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med 2015; 373: 726-36. PubMedPMC
- 29. Mazieres J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol 2013; 31: 1997-2003. ArticlePubMed
- 30. Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov 2015; 5: 842-9. ArticlePubMedPMCPDF
- 31. Awad MM, Oxnard GR, Jackman DM, et al. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J Clin Oncol 2016; 34: 721-30. ArticlePubMed
- 32. D’Angelo SP, Pietanza MC, Johnson ML, et al. Incidence of EGFR exon 19 deletions and L858R in tumor specimens from men and cigarette smokers with lung adenocarcinomas. J Clin Oncol 2011; 29: 2066-70. ArticlePubMedPMC
- 33. Sun PL, Seol H, Lee HJ, et al. High incidence of EGFR mutations in Korean men smokers with no intratumoral heterogeneity of lung adenocarcinomas: correlation with histologic subtypes, EGFR/TTF-1 expressions, and clinical features. J Thorac Oncol 2012; 7: 323-30. ArticlePubMed
- 34. Pirker R, Herth FJ, Kerr KM, et al. Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol 2010; 5: 1706-13. ArticlePubMed
- 35. Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011; 6: 244-85. PubMedPMC
- 36. Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol 2013; 8: 823-59. ArticlePubMedPMC
- 37. Noh S, Shim H. Optimal combination of immunohistochemical markers for subclassification of non-small cell lung carcinomas: a tissue microarray study of poorly differentiated areas. Lung Cancer 2012; 76: 51-5. ArticlePubMed
- 38. Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res 2007; 13: 4954-5. ArticlePubMedPDF
- 39. Lee HJ, Xu X, Kim H, et al. Comparison of direct sequencing, PNA clamping-real time polymerase chain reaction, and pyrosequencing methods for the detection of EGFR mutations in non-small cell lung carcinoma and the correlation with clinical responses to EGFR tyrosine kinase inhibitor treatment. Korean J Pathol 2013; 47: 52-60. ArticlePubMedPMC
- 40. Kim HJ, Lee KY, Kim YC, et al. Detection and comparison of peptide nucleic acid-mediated real-time polymerase chain reaction clamping and direct gene sequencing for epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Lung Cancer 2012; 75: 321-5. ArticlePubMed
- 41. Han HS, Lim SN, An JY, et al. Detection of EGFR mutation status in lung adenocarcinoma specimens with different proportions of tumor cells using two methods of differential sensitivity. J Thorac Oncol 2012; 7: 355-64. ArticlePubMed
- 42. Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015; 5: 850-9. ArticlePubMedPDF
- 43. Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013; 31: 1023-31. PubMedPMC
- 44. Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014; 20: 1479-84. ArticlePubMedPDF
- 45. Rekhtman N, Brandt SM, Sigel CS, et al. Suitability of thoracic cytology for new therapeutic paradigms in non-small cell lung carcinoma: high accuracy of tumor subtyping and feasibility of EGFR and KRAS molecular testing. J Thorac Oncol 2011; 6: 451-8. ArticlePubMed
- 46. Navani N, Brown JM, Nankivell M, et al. Suitability of endobronchial ultrasound-guided transbronchial needle aspiration specimens for subtyping and genotyping of non-small cell lung cancer: a multicenter study of 774 patients. Am J Respir Crit Care Med 2012; 185: 1316-22. ArticlePubMedPMC
- 47. Sun PL, Jin Y, Kim H, Lee CT, Jheon S, Chung JH. High concordance of EGFR mutation status between histologic and corresponding cytologic specimens of lung adenocarcinomas. Cancer Cytopathol 2013; 121: 311-9. ArticlePubMed
- 48. Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol 2011; 29: 2972-7. ArticlePubMed
- 49. Chung JH, Choe G, Jheon S, et al. Epidermal growth factor receptor mutation and pathologic-radiologic correlation between multiple lung nodules with ground-glass opacity differentiates multicentric origin from intrapulmonary spread. J Thorac Oncol 2009; 4: 1490-5. ArticlePubMed
- 50. Eberhard DA, Giaccone G, Johnson BE; Non-Small-Cell Lung Cancer Working Group. Biomarkers of response to epidermal growth factor receptor inhibitors in Non-Small-Cell Lung Cancer Working Group: standardization for use in the clinical trial setting. J Clin Oncol 2008; 26: 983-94. ArticlePubMed
- 51. Nicholson AG, Gonzalez D, Shah P, et al. Refining the diagnosis and EGFR status of non-small cell lung carcinoma in biopsy and cytologic material, using a panel of mucin staining, TTF-1, cytokeratin 5/6, and P63, and EGFR mutation analysis. J Thorac Oncol 2010; 5: 436-41. ArticlePubMed
- 52. Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol 2016; 2: 1014-22. ArticlePubMedPMC
- 53. Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol 2016; 34: 3375-82. ArticlePubMedPMC
- 54. Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer 2013; 13: 772-87. ArticlePubMedPMCPDF
- 55. Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun 2014; 5: 4846.ArticlePubMedPMCPDF
- 56. Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014; 371: 2167-77. ArticlePubMed
- 57. Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014; 371: 1963-71. PubMedPMC
- 58. Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012; 18: 375-7. ArticlePubMedPMCPDF
- 59. Wang R, Hu H, Pan Y, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol 2012; 30: 4352-9. ArticlePubMed
- 60. Drilon A, Wang L, Hasanovic A, et al. Response to cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov 2013; 3: 630-5. ArticlePubMedPMCPDF
- 61. Yoh K, Seto T, Satouchi M, et al. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med 2017; 5: 42-50. ArticlePubMed
- 62. Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med 2013; 19: 1469-72. ArticlePubMedPMCPDF
- 63. Wang R, Wang L, Li Y, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res 2014; 20: 4107-14. ArticlePubMedPDF
- 64. Nakaoku T, Tsuta K, Ichikawa H, et al. Druggable oncogene fusions in invasive mucinous lung adenocarcinoma. Clin Cancer Res 2014; 20: 3087-93. ArticlePubMedPMCPDF
- 65. Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013; 3: 636-47. ArticlePubMedPMCPDF
- 66. Paik JH, Choe G, Kim H, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: correlation with fluorescence in situ hybridization. J Thorac Oncol 2011; 6: 466-72. ArticlePubMed
- 67. Cha YJ, Lee JS, Kim HR, et al. Screening of ROS1 rearrangements in lung adenocarcinoma by immunohistochemistry and comparison with ALK rearrangements. PLoS One 2014; 9: e103333. Article
- 68. Lee SE, Lee B, Hong M, et al. Comprehensive analysis of RET and ROS1 rearrangement in lung adenocarcinoma. Mod Pathol 2015; 28: 468-79. ArticlePubMedPDF
- 69. Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res 2010; 16: 1561-71. ArticlePubMedPMCPDF
- 70. Conklin CM, Craddock KJ, Have C, Laskin J, Couture C, Ionescu DN. Immunohistochemistry is a reliable screening tool for identification of ALK rearrangement in non-small-cell lung carcinoma and is antibody dependent. J Thorac Oncol 2013; 8: 45-51. ArticlePMC
- 71. Soda M, Isobe K, Inoue A, et al. A prospective PCR-based screening for the EML4-ALK oncogene in non-small cell lung cancer. Clin Cancer Res 2012; 18: 5682-9. ArticlePubMedPDF
- 72. Betz BL, Dixon CA, Weigelin HC, Knoepp SM, Roh MH. The use of stained cytologic direct smears for ALK gene rearrangement analysis of lung adenocarcinoma. Cancer Cytopathol 2013; 121: 489-99. ArticlePubMed
- 73. Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010; 2: 62ra93.ArticlePubMedPMC
- 74. Kim HR, Kim DJ, Kang DR, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol 2013; 31: 731-7. ArticlePubMed
- 75. Dutt A, Ramos AH, Hammerman PS, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One 2011; 6: e20351. Article
- 76. Lim SH, Sun JM, Choi YL, et al. Efficacy and safety of dovitinib in pretreated patients with advanced squamous non-small cell lung cancer with FGFR1 amplification: a single-arm, phase 2 study. Cancer 2016; 122: 3024-31. ArticlePubMed
- 77. Wynes MW, Hinz TK, Gao D, et al. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res 2014; 20: 3299-309. ArticlePubMedPMCPDF
- 78. Pearson A, Smyth E, Babina IS, et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov 2016; 6: 838-51. ArticlePubMedPMCPDF
- 79. Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst 2005; 97: 643-55. ArticlePubMed
- 80. Soh J, Okumura N, Lockwood WW, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One 2009; 4: e7464. Article
- 81. Lee Y, Shim HS, Park MS, et al. High EGFR gene copy number and skin rash as predictive markers for EGFR tyrosine kinase inhibitors in patients with advanced squamous cell lung carcinoma. Clin Cancer Res 2012; 18: 1760-8. ArticlePubMedPDF
- 82. Schildhaus HU, Schultheis AM, Rüschoff J, et al. MET amplification status in therapy-naive adeno- and squamous cell carcinomas of the lung. Clin Cancer Res 2015; 21: 907-15. PubMed
- 83. Ou SH, Kwak EL, Siwak-Tapp C, et al. Activity of crizotinib (PF02341066), a dual mesenchymal-epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J Thorac Oncol 2011; 6: 942-6. ArticlePubMed
- 84. Zhang Y, Wang W, Wang Y, et al. Response to crizotinib observed in lung adenocarcinoma with MET copy number gain but without a high-level MET/CEP7 ratio, MET overexpression, orexon 14 splicing mutations. J Thorac Oncol 2016; 11: e59-62. ArticlePubMed
- 85. Mar N, Vredenburgh JJ, Wasser JS. Targeting HER2 in the treatment of non-small cell lung cancer. Lung Cancer 2015; 87: 220-5. ArticlePubMed
- 86. Takezawa K, Pirazzoli V, Arcila ME, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov 2012; 2: 922-33. ArticlePubMedPMCPDF
- 87. Schildhaus HU, Heukamp LC, Merkelbach-Bruse S, et al. Definition of a fluorescence in-situ hybridization score identifies high- and low-level FGFR1 amplification types in squamous cell lung cancer. Mod Pathol 2012; 25: 1473-80. ArticlePubMedPMCPDF
- 88. Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013; 31: 3997-4013. PubMed
- 89. Dziadziuszko R, Wynes MW, Singh S, et al. Correlation between MET gene copy number by silver in situ hybridization and protein expression by immunohistochemistry in non-small cell lung cancer. J Thorac Oncol 2012; 7: 340-7. ArticlePubMedPMC
- 90. Gadgeel SM, Chen W, Cote ML, et al. Fibroblast growth factor receptor 1 amplification in non-small cell lung cancer by quantitative real-time PCR. PLoS One 2013; 8: e79820. Article
- 91. Goke F, Franzen A, Menon R, et al. Rationale for treatment of metastatic squamous cell carcinoma of the lung using fibroblast growth factor receptor inhibitors. Chest 2012; 142: 1020-6. ArticlePubMed
REFERENCES
Figure & Data
References
Citations

- Association between PD-L1 expression with EGFR, ALK, and ROS1 driver oncogene mutations in non-small cell lung cancer
Dülger Onur, Yaylım İlhan, Öz Büge
Indian Journal of Pathology and Microbiology.2025; 68(1): 36. CrossRef - Enhancing Lung Cancer Care in Portugal: Bridging Gaps for Improved Patient Outcomes
Raquel Ramos, Conceição Souto Moura, Mariana Costa, Nuno Jorge Lamas, Renato Correia, Diogo Garcez, José Miguel Pereira, Carlos Sousa, Nuno Vale
Journal of Personalized Medicine.2024; 14(5): 446. CrossRef - Evolution of therapy for ALK-positive lung carcinomas: Application of third-generation ALK inhibitors in real clinical practice
A. F. Nasretdinov, A. V. Sultanbaev, Sh. I. Musin, K. V. Menshikov, R. T. Ayupov, A. A. Izmailov, G. A. Serebrennikov, V. E. Askarov, D. V. Feoktistov
Meditsinskiy sovet = Medical Council.2024; (10): 74. CrossRef - Cost-effectiveness of next-generation sequencing for advanced EGFR/ALK-negative non-small cell lung cancer
Dong-Won Kang, Sun-Kyeong Park, Sokbom Kang, Eui-Kyung Lee
Lung Cancer.2024; 197: 107970. CrossRef - miR-92a-3p regulates cisplatin-induced cancer cell death
Romain Larrue, Sandy Fellah, Nihad Boukrout, Corentin De Sousa, Julie Lemaire, Carolane Leboeuf, Marine Goujon, Michael Perrais, Bernard Mari, Christelle Cauffiez, Nicolas Pottier, Cynthia Van der Hauwaert
Cell Death & Disease.2023;[Epub] CrossRef - Diagnostic Approach of Lung Cancer: A Literature Review
Jesi Hana, Novia Nurul Faizah
Jurnal Respirasi.2023; 9(2): 141. CrossRef - Molecular Pathology of Lung Cancer
James J. Saller, Theresa A. Boyle
Cold Spring Harbor Perspectives in Medicine.2022; 12(3): a037812. CrossRef - Landscape of EGFR mutations in lung adenocarcinoma: a single institute experience with comparison of PANAMutyper testing and targeted next-generation sequencing
Jeonghyo Lee, Yeon Bi Han, Hyun Jung Kwon, Song Kook Lee, Hyojin Kim, Jin-Haeng Chung
Journal of Pathology and Translational Medicine.2022; 56(5): 249. CrossRef - Molecular biomarker testing for non–small cell lung cancer: consensus statement of the Korean Cardiopulmonary Pathology Study Group
Sunhee Chang, Hyo Sup Shim, Tae Jung Kim, Yoon-La Choi, Wan Seop Kim, Dong Hoon Shin, Lucia Kim, Heae Surng Park, Geon Kook Lee, Chang Hun Lee
Journal of Pathology and Translational Medicine.2021; 55(3): 181. CrossRef - TM4SF4 and LRRK2 Are Potential Therapeutic Targets in Lung and Breast Cancers through Outlier Analysis
Kyungsoo Jung, Joon-Seok Choi, Beom-Mo Koo, Yu Jin Kim, Ji-Young Song, Minjung Sung, Eun Sol Chang, Ka-Won Noh, Sungbin An, Mi-Sook Lee, Kyoung Song, Hannah Lee, Ryong Nam Kim, Young Kee Shin, Doo-Yi Oh, Yoon-La Choi
Cancer Research and Treatment.2021; 53(1): 9. CrossRef - The promises and challenges of early non‐small cell lung cancer detection: patient perceptions, low‐dose CT screening, bronchoscopy and biomarkers
Lukas Kalinke, Ricky Thakrar, Sam M. Janes
Molecular Oncology.2021; 15(10): 2544. CrossRef - Cost-effectiveness analyses of targeted therapy and immunotherapy for advanced non-small cell lung cancer in the United States: a systematic review
Anthony Yu, Eva Huang, Momoka Abe, Kang An, Sun-Kyeong Park, Chanhyun Park
Expert Review of Pharmacoeconomics & Outcomes Research.2021; 21(3): 381. CrossRef - The expanding capability and clinical relevance of molecular diagnostic technology to identify and evaluate EGFR mutations in advanced/metastatic NSCLC
Parth Shah, Jacob Sands, Nicola Normanno
Lung Cancer.2021; 160: 118. CrossRef - Testing for EGFR Mutations and ALK Rearrangements in Advanced Non-Small-Cell Lung Cancer: Considerations for Countries in Emerging Markets
Mercedes L Dalurzo, Alejandro Avilés-Salas, Fernando Augusto Soares, Yingyong Hou, Yuan Li, Anna Stroganova, Büge Öz, Arif Abdillah, Hui Wan, Yoon-La Choi
OncoTargets and Therapy.2021; Volume 14: 4671. CrossRef - Treatment of Patients With Non–Small-Cell Lung Cancer Harboring Rare Oncogenic Mutations
Melina E. Marmarelis, Corey J. Langer
Clinical Lung Cancer.2020; 21(5): 395. CrossRef - Detection of Targetable Genetic Alterations in Korean Lung Cancer Patients: A Comparison Study of Single-Gene Assays and Targeted Next-Generation Sequencing
Eunhyang Park, Hyo Sup Shim
Cancer Research and Treatment.2020; 52(2): 543. CrossRef - High prevalence of ROS1 gene rearrangement detected by FISH in EGFR and ALK negative lung adenocarcinoma
Yuyin Xu, Heng Chang, Lijing Wu, Xin Zhang, Ling Zhang, Jing Zhang, Yuan Li, Lei Shen, Xiaoli Zhu, Xiaoyan Zhou, Qianming Bai
Experimental and Molecular Pathology.2020; 117: 104548. CrossRef - An All-In-One Transcriptome-Based Assay to Identify Therapy-Guiding Genomic Aberrations in Nonsmall Cell Lung Cancer Patients
Jiacong Wei, Anna A. Rybczynska, Pei Meng, Martijn Terpstra, Ali Saber, Jantine Sietzema, Wim Timens, Ed Schuuring, T. Jeroen N. Hiltermann, Harry. J.M. Groen, Anthonie van der Wekken, Anke van den Berg, Klaas Kok
Cancers.2020; 12(10): 2843. CrossRef - Immunotherapy in EGFR-Mutant and ALK-Positive Lung Cancer
Alexander Gavralidis, Justin F. Gainor
The Cancer Journal.2020; 26(6): 517. CrossRef - Role of Immunocytochemistry in the Cytological Diagnosis of Pulmonary Tumors
Jasna Metovic, Luisella Righi, Luisa Delsedime, Marco Volante, Mauro Papotti
Acta Cytologica.2020; 64(1-2): 16. CrossRef - Molecular Diagnostic Assays and Clinicopathologic Implications of MET Exon 14 Skipping Mutation in Non–small-cell Lung Cancer
Eun Kyung Kim, Kyung A. Kim, Chang Young Lee, Sangwoo Kim, Sunhee Chang, Byoung Chul Cho, Hyo Sup Shim
Clinical Lung Cancer.2019; 20(1): e123. CrossRef - PD‐L1 expression in ROS1‐rearranged non‐small cell lung cancer: A study using simultaneous genotypic screening of EGFR, ALK, and ROS1
Jongmin Lee, Chan Kwon Park, Hyoung‐Kyu Yoon, Young Jo Sa, In Sook Woo, Hyo Rim Kim, Sue Youn Kim, Tae‐Jung Kim
Thoracic Cancer.2019; 10(1): 103. CrossRef - Human Leukocyte Antigen Class I and Programmed Death-Ligand 1 Coexpression Is an Independent Poor Prognostic Factor in Adenocarcinoma of the Lung
Yeon Bi Han, Hyun Jung Kwon, Soo Young Park, Eun-Sun Kim, Hyojin Kim, Jin-Haeng Chung
Journal of Pathology and Translational Medicine.2019; 53(2): 86. CrossRef - Molecular testing for advanced non-small cell lung cancer in Malaysia: Consensus statement from the College of Pathologists, Academy of Medicine Malaysia, the Malaysian Thoracic Society, and the Malaysian Oncological Society
Pathmanathan Rajadurai, Phaik Leng Cheah, Soon Hin How, Chong Kin Liam, Muhammad Azrif Ahmad Annuar, Norhayati Omar, Noriah Othman, Nurhayati Mohd Marzuki, Yong Kek Pang, Ros Suzanna Ahmad Bustamam, Lye Mun Tho
Lung Cancer.2019; 136: 65. CrossRef - Somatic mutations and immune checkpoint biomarkers
Brielle A. Parris, Eloise Shaw, Brendan Pang, Richie Soong, Kwun Fong, Ross A. Soo
Respirology.2019; 24(3): 215. CrossRef - Adverse Event Management in Patients with BRAF V600E-Mutant Non-Small Cell Lung Cancer Treated with Dabrafenib plus Trametinib
Anna Chalmers, Laura Cannon, Wallace Akerley
The Oncologist.2019; 24(7): 963. CrossRef - Genetic and clinicopathologic characteristics of lung adenocarcinoma with tumor spread through air spaces
Jae Seok Lee, Eun Kyung Kim, Moonsik Kim, Hyo Sup Shim
Lung Cancer.2018; 123: 121. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure


Fig. 1.
Fig. 2.
| Gene | Representative subtypes or variants | Frequency | Targeted agents |
|---|---|---|---|
| Mutations | |||
| EGFR | Exon 19 deletion, Exon 21 L858R, Exon 20 T790M | 40%–50% in ADCs |
Gefitinib, erlotinib, afatinib, osimertinib |
| 10%–20% in ADCs |
|||
| KRAS | G12X, G13X, G61X | 5%–10% in ADCs |
MEK inhibitors |
| 20%–30% in ADCs |
|||
| BRAF | V600E | 1%–4% in ADCs | Vemurafenib, dabrafenib, |
| HER2 | p.A775 G776insYVMA in exon 20 | 1%–2% in ADCs | Trastuzumab, afatinib |
| MET | Splice site mutations around or in exon 14 | 3%–4% in ADCs | Crizotinib, cabozantinib |
| Gene fusions | |||
| ALK | EML4-ALK, TGF-ALK, KIF5B-ALK | 5% in ADCs | Crizotinib, ceritinib, alectinib |
| ROS1 | CD74-ROS1, EZR-ROS1, SLC34A2-ROS1, SDC4-ROS1 | 1% in ADCs | Crizotinib, ceritinib |
| RET | KIF5B-RET, CCDC6-RET | 1% in ADCs | Cabozantinib, vandetanib, alectinib |
| NTRK1 | MPRIP-NTRK1 and CD74-NTRK1, TPM3-NTRK1 | < 1% in ADCs | Entrectinib |
| FGFR1/3 | FGFR3-TACC3, BAG4-FGFR1 | 1% in NSCLCs | FGFR inhibitor |
| NRG1 | CD74-NRG1, SLC3A2-NRG1, VAMP2-NRG1 | 7% in mucinous ADCs | NA |
| Amplifications | |||
| FGFR1 | Gene amplification | 13%–22% in SQCs | FGFR inhibitor |
| EGFR | Gene amplification | 8%–9% in SQCs, | EGFR inhibitor |
| MET | Gene amplification | 2%–4% in ADCs | Crizotinib |
| HER2 | Gene amplification | 1%–2% in ADCs | Trastuzumab, afatinib |
| Category | Mutation | Gene rearrangement | Amplification |
|---|---|---|---|
| DNA | Direct sequencing | FISH | FISH |
| PCR-based methods | NGS | qPCR | |
| NGS | NGS | ||
| RNA | RT-PCR (fusion transcript) | Real-time PCR (mRNA overexpression) | |
| NGS | |||
| Protein | IHC (mutation-specific antibody) | IHC (protein expression) | IHC (protein overexpression) |
| Recommendation | |||
|---|---|---|---|
| Indications | Mutations | ||
| Do: adenocarcinoma, large cell carcinoma, NSCLC-NOS | |||
| Consider: at a young age, never smokers or small biopsy samples or mixed histological features | |||
| All types if clinically indicated | |||
| Gene rearrangements | |||
| Do: adenocarcinoma, large cell carcinoma, NSCLC-NOS for most gene fusions | |||
| Consider: at a young age, never smokers or small biopsy samples or mixed histological features | |||
| Squamous cell carcinoma for FGFR fusions | |||
| All types if clinically indicated | |||
| Amplifications | |||
| All types if clinically indicated | |||
| Squamous cell carcinoma for FGFR1 amplification | |||
| Adenocarcinoma, NSCLC-NOS for MET or HER2 amplifications | |||
| Method | An appropriate method should be selected according to genetic alterations. | ||
| The pathologist should consider the pros and cons of each method. | |||
| Type of specimen | Histological and cytological samples are both acceptable. | ||
| Either a primary tumor or a metastatic lesion is equally suitable. | |||
| In cases of multiple, synchronous primary lung adenocarcinomas, each tumor may be tested. | |||
| Specimen requirements | The presence of tumor cells must be verified by a pathologist. | ||
| High percentage (ideally more than 50%) of tumor cells for direct sequencing | |||
| Lower percentage acceptable for methods with higher sensitivity | |||
| A minimum of 50-100 assessable tumor cells are required for a FISH assay. | |||
| Reporting | Patients and sample information, material used for analysis, type of method, results of the test, comments, names of the testing technician and corresponding pathologist | ||
| Validation of test | New methods must be approved by the Korean government. | ||
| Analytical and clinical validation procedures should be conducted when the testing is set up in the laboratory. | |||
| A combination of more than one method may be useful in equivocal cases. | |||
| Quality assurance | Quality assurance program (internal or external quality control) should be implemented. | ||
ADC, adenocarcinoma; NSCLC, non-small cell lung carcinoma; FGFR, fibroblast growth factor receptor; NA, not available; SQC, squamous cell carcinoma; EGFR, epidermal growth factor receptor. Asian populations; Western populations.
PCR, polymerase chain reaction; NGS, next-generation sequencing; FISH, fluorescence
NSCLC-NOS, non-small cell lung carcinoma not otherwise specified; FGFR, fibroblast growth factor receptor; FISH, fluorescence

 E-submission
E-submission