Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 49(5); 2015 > Article
-
Original Article
SALL4 Expression in Hepatocellular Carcinomas Is Associated with EpCAM-Positivity and a Poor Prognosis - Hyunjin Park, Hyejung Lee, An Na Seo1, Jai Young Cho2, Young Rok Choi2, Yoo-Seok Yoon2, Ho-Seong Han2, Young Nyun Park3, Haeryoung Kim
-
Journal of Pathology and Translational Medicine 2015;49(5):373-381.
DOI: https://doi.org/10.4132/jptm.2015.07.09
Published online: August 10, 2015
Department of Pathology, Seoul National University Bundang Hospital, Seoul National University College of Medicine, Seoul, Korea
1Department of Pathology, Kyungpook National University Medical Center, Kyungpook National University School of Medicine, Daegu, Korea
2Department of Surgery, Seoul National University Bundang Hospital, Seoul National University College of Medicine, Seoul, Korea
3Department of Pathology, Yonsei University College of Medicine, Seoul, Korea
-
Corresponding Author Haeryoung Kim, MD, PhD Department of Pathology, Seoul National University Bundang Hospital, Seoul National University College of Medicine, 82 Gumi-ro 173beon-gil, Bundang-gu, Seongnam 13620, Korea Tel: +82-31-787-7715 Fax: +82-31-787-4012 E-mail: haeryoung.kim@snu.ac.kr
Corresponding Author Young Nyun Park, MD, PhD Department of Pathology, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, Korea Tel: +82-2-2228-1678 Fax: +82-2-362-0860 E-mail: young0608@yuhs.ac
© 2015 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background:
- There is increasing interest in hepatocellular carcinomas (HCC) expressing “stemness”-related markers, as they have been associated with aggressive behavior and poor prognosis. In this study, we investigated the usefulness of Sal-like protein 4 (SALL4), a recently proposed candidate marker of “stemness.”
-
Methods:
- Immunohistochemical stains were performed for SALL4, K19, and epithelial cellular adhesion molecule (EpCAM) on tissue microarrays constructed from 190 surgically resected HCCs, and the results were correlated with the clinicopathological features and patient survival data.
-
Results:
- Nuclear SALL4 expression was observed in 39/190 HCCs (20.5%), while K19 and EpCAM were expressed in 30 (15.9%) and 92 (48.7%) HCCs, respectively. The nuclear expression was generally weak, punctate or clumped. SALL4 expression was significantly associated with a poor overall survival compared to SALL4-negative HCCs (p = .014) compared to SALL4-negative HCCs. On multivariate analysis adjusted for tumor size, multiplicity, vascular invasion, and pathological tumor stage, SALL4 remained as a significant independent predictor of decreased overall survival (p= .004). SALL4 expression was positively correlated with EpCAM expression (p = .013) but not with K19 expression. HCCs that expressed both SALL4 and EpCAM were associated with significantly decreased overall survival, compared to those cases which were negative for both of these markers (p = .031).
-
Conclusions:
- Although SALL4 expression was not significantly correlated with other clinicopathological parameters suggestive of tumor aggressiveness, SALL4 expression was an independent predictor of poor overall survival in human HCCs, and was also positively correlated with EpCAM expression.
- Case selection and review
- This study was approved by the Institutional Review Board of Seoul National University Bundang Hospital (B-1502-286-302). One hundred and ninety consecutive cases of surgically resected or explanted HCCs were enrolled in this study, and were retrieved from the surgical pathology files of Seoul National University Bundang Hospital from May 2003 to April 2010. The clinicopathological characteristics of the cases are summarized in Table 1. The electronic medical records, surgical pathology reports, and the hematoxylin and eosin-stained slides were reviewed for each case, and the clinicopathological variables noted included tumor size, multiplicity (including intrahepatic metastasis/satellite nodules and multicentric occurrences), histological differentiation according to the Edmondson-Steiner grade (the highest grade in tumors with heterogeneity), the presence of microvascular or major vascular invasion, the patient demographics (age and sex), and the presence of an underlying etiology (e.g., hepatitis B, C, alcohol). Follow-up data was also obtained from the medical records, including recurrences (including local recurrence and distant metastasis) and death. The median follow-up period was 52 months (range, 0 to 133 months). Overall survival was defined as thfe interval from initial surgical treatment for HCC to the date of death, and disease-free survival as the interval from initial treatment to local or distant recurrence.
- Tissue microarray construction and immunohistochemistry
- Tissue cores measuring 2 mm in diameter were sampled from formalin-fixed paraffin-embedded HCC tissues and arranged in recipient tissue array blocks using a trephine apparatus (Superbiochips Laboratories, Seoul, Korea). Two or three cores were sampled from each HCC, depending on the amount of histological heterogeneity present in the tumor. One core was sampled from the corresponding non-neoplastic liver for each case. Four μm-thick sections were obtained from the tissue microarray blocks and subjected to immunohistochemical staining for SALL4 (1:100, mouse monoclonal antibody, clone EE-30, Santa Cruz Technologies Inc., Santa Cruz, CA, USA), K19 (1:150, mouse monoclonal antibody, clone BA17, Dako, Glostrup, Denmark) and EpCAM (1:3,000, mouse monoclonal antibody, clone VU-1D9, Calbiochem, Darmstadt, Germany). Briefly, tissue sections were deparaffinized in xylene, rehydrated in graded alcohol, and antigen retrieval was performed using citrate buffer (pH 6.0) for 15 minutes. Sections were incubated with the primary antibodies at room temperature for 30 minutes, and then incubated with secondary antibodies (EnVision Detection System, Dako). Counterstaining was performed using Mayer’s hematoxylin and the stained slides were mounted.
- SALL4 was expressed in the tumor cell nuclei, and positivity for SALL4 expression was defined as nuclear staining for the protein in more than 10% of the tumor cells after calculating the SALL4-labeling index (a ratio of positive nuclei/total nuclei under the 400× field magnification) with the help of the Image J software (downloaded from http://imagej.nih.gov/ij). A testicular seminoma tissue sample was used as a positive control for SALL4. Membranous and/or cytoplasmic staining in the tumor cells were counted as positive for EpCAM and K19.
- Statistical analysis
- All statistical analyses were conducted using IBM SPSS ver. 21 (IBM Corp., Armonk, NY, USA). Chi-square, Fisher’s exact and t tests were performed as deemed appropriate. Univariable analyses for overall survival and disease-free survival were performed using the Kaplan-Meier method and log-rank tests. Statistically significant variables from the univariable analysis were entered into the multivariable analyses using the Cox proportional hazard method. Statistical significance was defined as p<.05.
MATERIALS AND METHODS
- SALL4, K19, and EpCAM expression in HCC
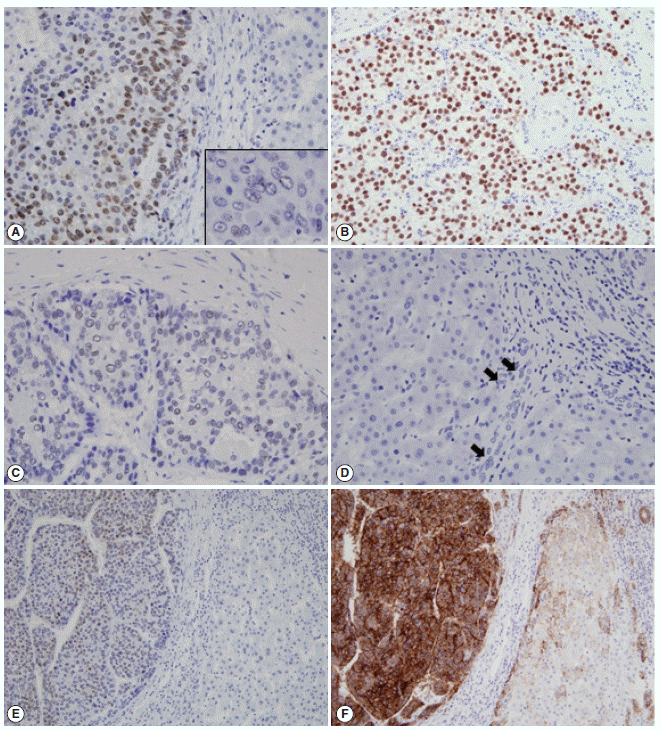
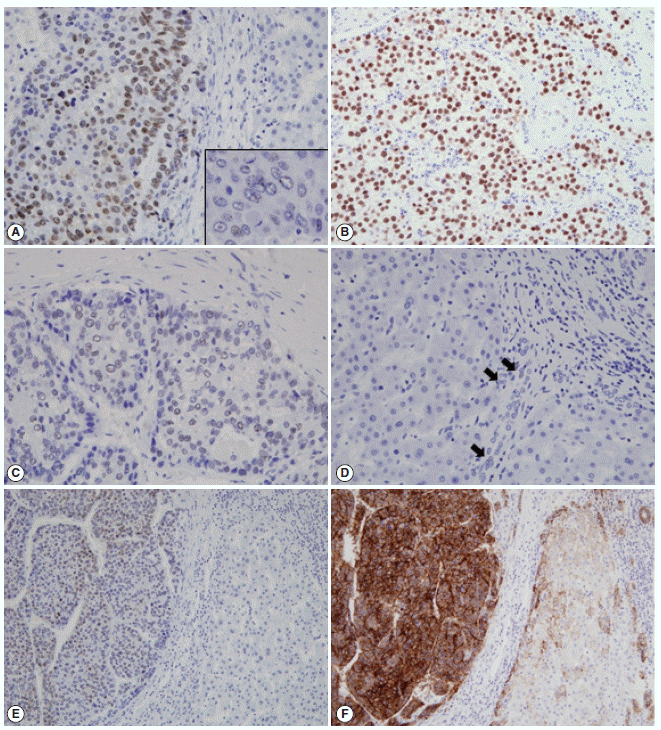
- The immunohistochemical stain results and the expression frequencies of SALL4, K19, and EpCAM are summarized in Figs. 1 and 2, respectively. The SALL4 nuclear labeling index ranged from 0% to 78.9% (mean±standard deviation, 8.9±19.2%) in the 190 HCCs, and 39 cases (20.5%) were deemed positive for SALL4 expression (labeling index ≥10%). Although a definite nuclear staining was present in the tumor cells, the intensity of staining was generally not as strong as in the positive control tissue (testicular seminoma) (Fig. 1A–C). In some areas, the nuclear staining was punctate and only identifiable at higher power magnification. SALL4 positivity was generally uniformly distributed in the HCCs without predilections for a particular morphological tumor cell: some SALL4-positive tumor cells were small with increased nuclear/cytoplasmic ratios reminiscent of the stem/progenitor cell phenotype, while others were more typical HCCs with various degrees of differentiation. In the adjacent non-neoplastic liver, the hepatocytes and bile ducts did not stain for SALL4, and interestingly, SALL4 positivity was seen in occasional ductular reactions, although the staining was very faint and barely visible (Fig. 1D).
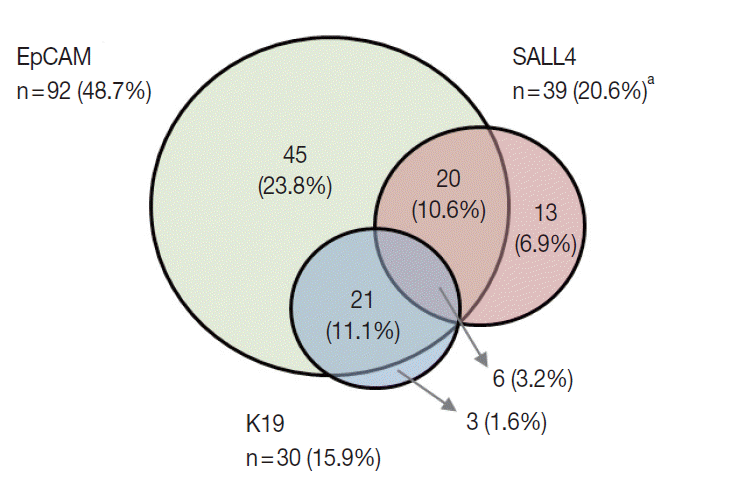
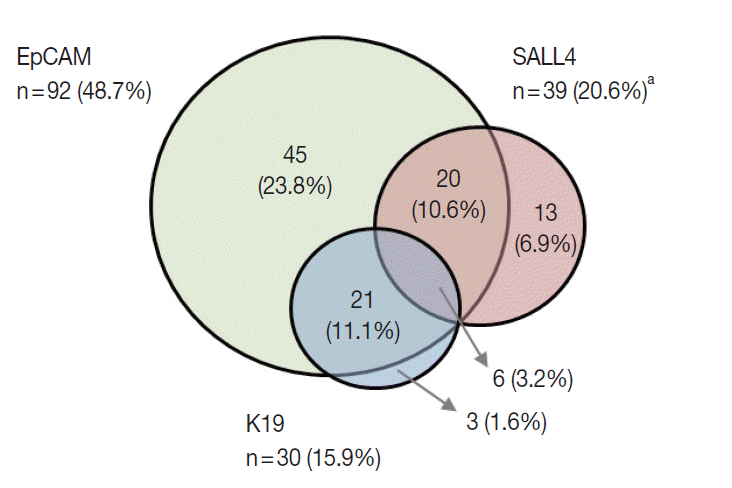
- K19 and EpCAM expression data were available for 189 cases (due to tissue core loss in 1 case). K19 and EpCAM were expressed in 30 (15.9%) and 92 (48.7%) cases, respectively, in the typical membranous/cytoplasmic pattern, and they were also expressed in the bile ducts and ductular reactions in the non-neoplastic livers. A significant positive correlation was seen between EpCAM and K19 expression: K19 positivity was seen in 27 out of 92 EpCAM-positive HCCs (29.3%) compared to 3 out of 97 EpCAM negative HCCs (3.1%) (p<.001). SALL4 expression was more frequently seen in EpCAM-positive HCCs (26/92, 28.3%) compared to EpCAM-negative HCCs (13/97, 13.4%) (p=.013) (Fig. 1E, F). However, there was no significant correlation between SALL4 and K19 expression: only 3.2% of the HCCs demonstrated positivity for both SALL4 and K19 (Fig. 2). The relationships between the expression of SALL4, K19, and EpCAM in HCCs are summarized in a Venn diagram in Fig. 2.
- Correlation with clinicopathological variables and survival
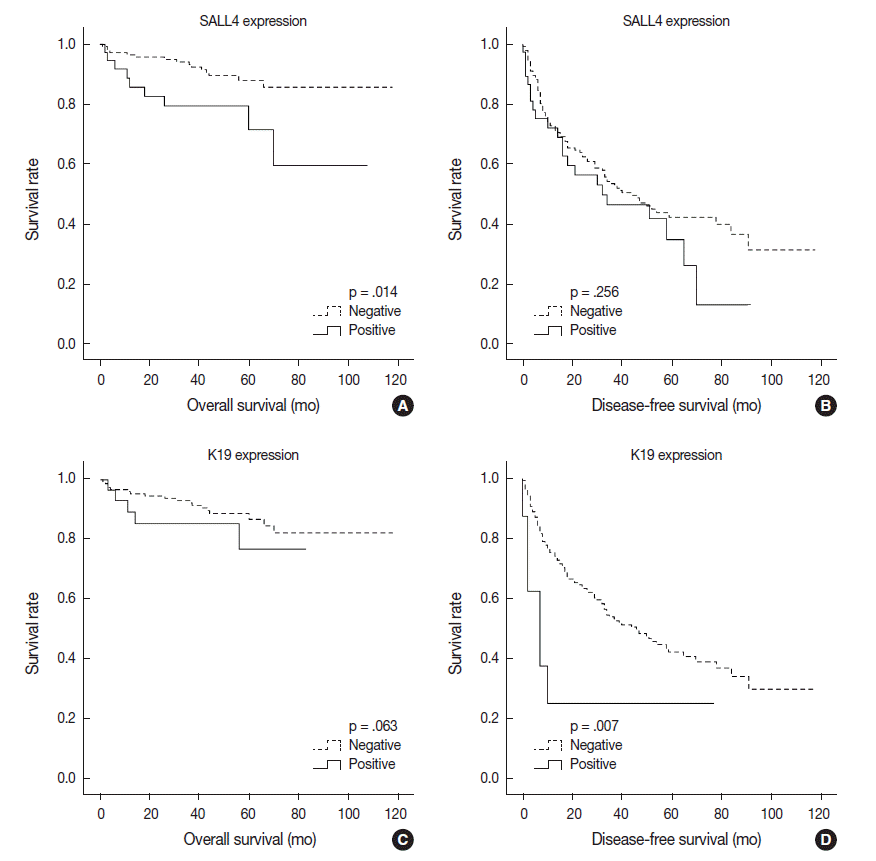
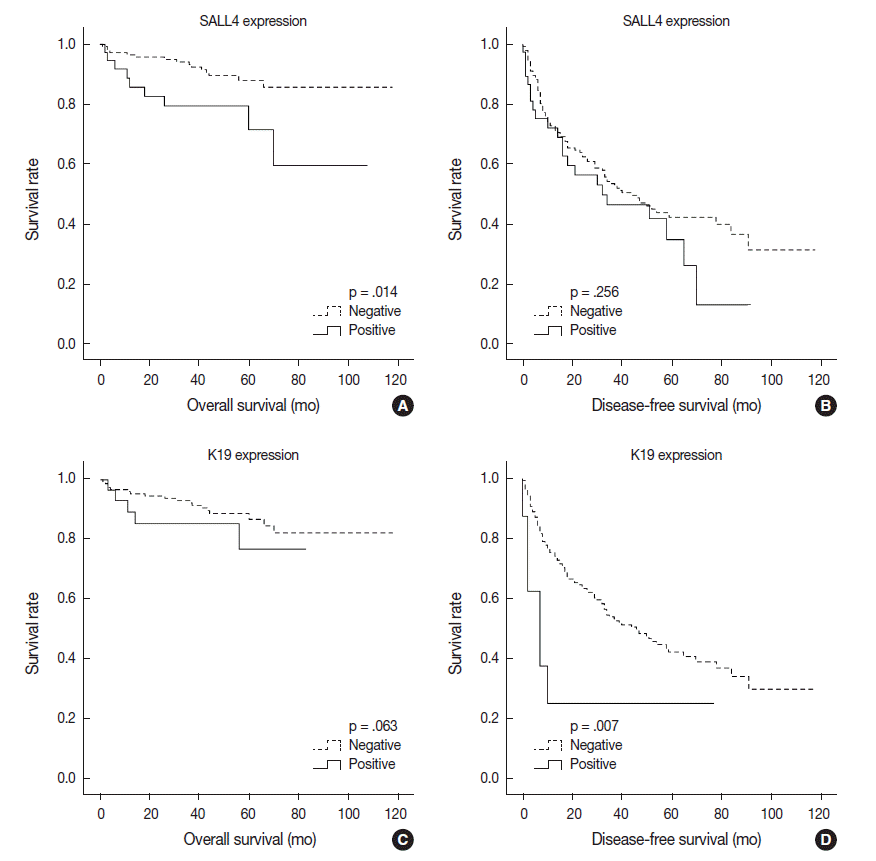
- Univariable analysis demonstrated that SALL4 expression was significantly associated with reduced overall survival (p=.014) (Fig. 3A). In addition to SALL4 expression, larger tumor size (>3 cm, p=.045), multiplicity of tumor (p=.001), major vascular invasion (p=.046) and higher T stage (stage 3 or 4, p<.001) were significantly associated with decreased overall survival. In comparison, SALL4 expression was not significantly associated with disease-free survival (Fig. 3B), while larger size (>3 cm, p=.009), multiplicity (p=.002) and higher T stage (stage 3 or 4, p=.019) were significantly associated with reduced disease-free survival. HCCs with K19 expression had a tendency for decreased overall survival, although this result was not statistically significant (p=.063), while disease-free survival was significantly decreased for K19 expressing HCCs (p=.001) (Fig. 3C, D). EpCAM expression was associated with a tendency for decreased overall survival (p=.053); however, it was not associated with disease-free survival.
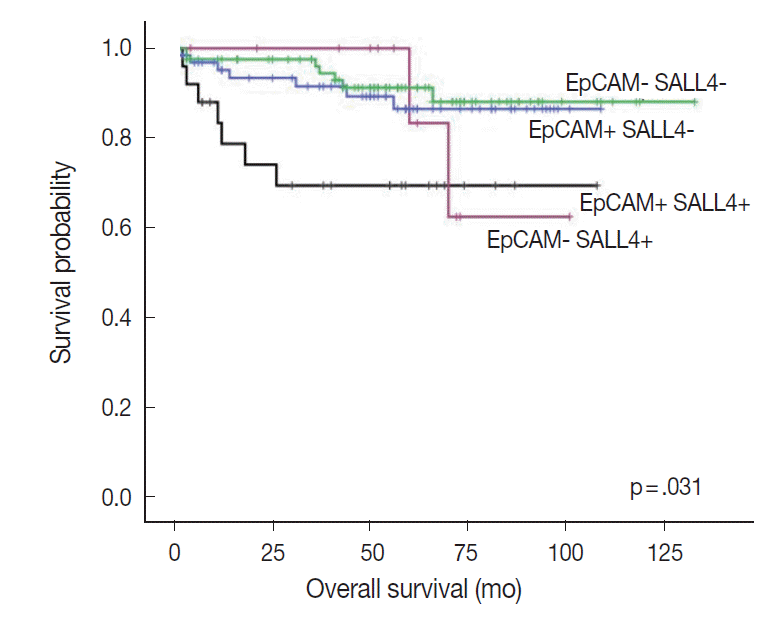
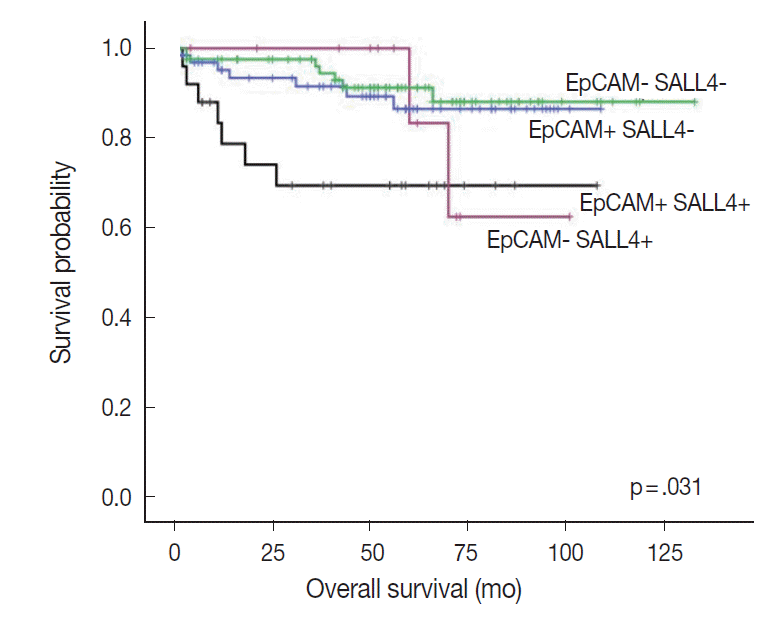
- Interestingly, when we performed the survival analysis after combining EpCAM and SALL4 expression status, HCCs that expressed both SALL4 and EpCAM were associated with significantly decreased overall survival, compared to those cases which were negative for both these markers (p=.031) (Fig. 4). In addition, while EpCAM-negative HCCs were associated with a relatively favorable outcome compared to EpCAM-positive HCCs during the earlier follow-up period (<5 years), EpCAM-negative HCCs with SALL4-positivity showed an abrupt decrease in overall survival after 5 years of follow-up.
- On multivariable analysis, SALL4 (p=.004), multiplicity (p=.016) and higher T stage (p=.004) remained as significant independent predictors of decreased overall survival (Table 2). For disease-free survival, larger tumor size (>3 cm, p=.019), and multiplicity (p=.004) were independent prognostic factors.
- SALL4 expression was not significantly associated with other clinicopathological parameters of tumor aggressiveness, such as presence of vascular invasion, larger tumors, and multiplicity (Table 3). On the other hand, EpCAM expression in HCCs was more frequently associated with poor histological differentiation (p=.019), microvascular invasion (p=.011), younger age (p=.020), B-viral etiology (p=.010), and high serum α-fetoprotein (AFP) levels (p=.023), and K19 expression was more frequently associated with higher serum AFP levels (p=.032), major vascular invasion (p=.010), and higher pathological T stage (3 or 4, p=.011). Increased tumor size was also more frequent in K19-positive HCCs, although not statistically significant (p=.076).
RESULTS
- HCCs with “stemness”-related marker expression—those that have the histomorphological features of conventional HCCs but express markers related to stemness on immunohistochemistry—have been receiving increasing interest over the past several years, as there is accumulating evidence that they are associated with aggressive behavior and poor prognosis, compared to conventional HCCs that do not express these markers [5,6]. The more frequently discussed markers of stemness in HCCs include K19, EpCAM, and CD133; however, it is still uncertain which marker best represents stemness in HCCs, and there is still a growing number of putative markers of stemness in the literature, including the recently proposed SALL4, which need further validation [10]. We were interested to see whether SALL4 expression in HCCs is indeed associated with the expression of other more established markers of stemness (K19 and EpCAM) and whether it has clinicopathological implications in our cohort of resected HCCs. Interestingly, we found SALL4 expression in 20.5% of HCCs, that SALL4 expression is positively correlated with EpCAM expression, and also that it has prognostic implications.
- SALL4 is a zinc finger transcriptional activator located on chromosome 20q13.13-13.2 that is required for the maintenance of pluripotent embryonic stem cells, potentially through interactions with Oct3/4, Sox2, and Nanog [12-14]. Mutation in SALL4 results in the Okihiro syndrome, a rare autosomal dominant disorder characterized by multiple organ defects [14]. SALL4 has been found to be expressed in leukemias and hematopoietic stem cells, and also in solid tumors such as germ cell tumors and AFP-producing gastric cancers, suggesting that SALL4 may be a marker of embryonic stem cells and also cancer stem cells [15-18]. In the fetal murine liver, SALL4 has been shown to be expressed in hepatic stem/progenitor cells, but the expression diminishes gradually during development and is not seen in adult hepatocytes [19]. These findings suggest that SALL4 may be a good marker of hepatic stem/progenitor cells and HCC cancer stem cells.
- Over the past two years, there has been increasing interest in SALL4 as a potential marker for identifying HCCs with features of “stemness,” aggressive behavior, and poor prognosis. Oikawa et al. [9] demonstrated a decreased overall survival in their profiling analysis of 110 HCCs (from the microarray dataset published by Lee et al. [1]), and also showed that the overexpression of SALL4 in liver cancer cells induced the expression of K19 and EpCAM, suggesting that SALL4 may be a marker of stem cells. Zeng et al. [11] demonstrated a decreased recurrence-free survival in SALL4-positive HCCs, and SALL4 activation in cell lines resulted in the up-regulation of hepatic stem cell markers KRT19, EPCAM, and CD44. Another extensive study showed a poor overall survival in SALL4-positive HCCs in two independent cohorts (Singapore, n=179; Hong Kong, n=228), in addition to an enrichment of progenitor-like gene signatures and overexpression of proliferative and metastatic genes on gene expression analysis [10]. In another recent analysis of Western HCCs, higher tumor grade, more frequent lymphovascular invasion and shorter recurrence-free and overall survivals were seen in SALL4-positive HCCs, although the SALL4-positivity was only seen very rarely (1.3%), suggesting differences in SALL4 expression status according to the etiology of HCC [20]. The expression frequency of SALL4 in HCC has in fact varied widely in different reports, ranging from 1.3% to 85% [9-11,20,21]. It is plausible that a higher prevalence of hepatitis B virus infection may account for the higher frequency of SALL4-positivity in Eastern HCCs; interestingly, SALL4-positivity has been shown to be more frequently associated with hepatitis B virus infection [11]. In addition, the differences in the antibodies, immunohistochemistry protocols, and the interpretation methods for defining “SALL4-positivity” are also likely to account for the wide range of SALL4-positivity reported in the literature.
- As it has been previously demonstrated that nuclear labeling for SALL4 in liver tissues is seen with high pH antigen retrieval and not as clearly with the conventional citrate buffer [9], we also stained the same tissue microarrays for SALL4 using a different antigen retrieval method (Tris-EDTA buffer; pH 9.0) to see if there were differences in the staining pattern or frequency; however, the results were identical (data not shown). Therefore, SALL4 immunostaining is feasible using the citrate buffer (pH 6.0) for antigen retrieval. We found that the nuclear staining for SALL4 in the testicular seminoma was very intense, diffuse and finely granular, in comparison to most SALL4-positive HCCs in which the nuclear staining was weaker and punctate or clumped, similar to a previous report [21]. In practice, when faced with the occasional diagnostic challenge of discriminating between HCC and yolk sac tumor—which may have similar morphological features and also positivity for AFP and glypican-3—SALL4 immunohistochemistry may play a role, as the strong and diffuse staining for SALL4 is rarely seen in HCCs [21]. However, as for the utility of SALL4 in HCCs as a prognostic marker and a marker of “stemness,” further validation in independent cohorts of HCC would be necessary, as the staining is not as intense, and there is likely to be interobserver variation in the interpretation of SALL4 positivity. Although we found a significantly decreased overall survival and a higher frequency of EpCAM positivity in SALL4-positive HCCs compared to SALL4-negative HCCs, we performed a semi-manual count of the SALL4-labeling index with the help of an image analysis software, which is not always a feasible method in routine pathology practice in comparison to the distinct cytoplasmic staining for K19 or EpCAM.
- Our finding of a positive correlation between SALL4 and EpCAM expression in HCCs, poor overall survival in SALL4-positive HCCs, and positive (albeit weak and sporadic) staining in the ductular reactions support the recent literature that SALL4 may be a useful marker of “stemness” in hepatic stem/progenitor cells and HCCs. However, in contrast to K19 or EpCAM, the nuclear staining is not as easily appreciable by immunohistochemistry, and other than being a prognostic indicator of poor overall survival, there were no significant correlations between SALL4 positivity and the clinicopathological parameters suggestive of aggressiveness. Therefore, while SALL4 is an independent prognostic factor for decreased overall survival in HCC patients and may be a potential marker for identifying the aggressive subgroup of HCCs with “stemness” features, further validation in larger HCC cohorts is required prior to further consideration of SALL4 as a good marker of “stemness” in liver specimens.
DISCUSSION
Acknowledgments
| Characteristic | No. (%) |
|---|---|
| Age (yr) | 58.1 ± 11.8 |
| Sex | |
| Male | 151 (79.5) |
| Female | 39 (20.1) |
| Etiology | |
| Hepatitis B | 136 (71.6) |
| Hepatitis C | 17 (8.9) |
| Alcohol | 8 (4.2) |
| Others | 30 (15.8) |
| Serum alpha-fetoprotein (IU/mL) | 1,634.6 ± 4,590.4 |
| Serum PIVKA-II (AU/mL) | 661.2 ± 1,528.9 |
| Multiplicity | |
| Absent | 106 (55.8) |
| Present | 31 (16.3) |
| Tumor size (cm)a | 4.6 ± 3.0 |
| Edmondson-Steiner grade | |
| I | 1 (0.5) |
| II | 47 (24.7) |
| III | 122 (64.2) |
| IV | 20 (10.5) |
| Microvascular invasion | |
| Absent | 116 (61.1) |
| Present | 74 (38.9) |
| Major vessel invasionb | |
| Absent | 168 (88.4) |
| Present | 22 (11.6) |
| Follow-up | |
| Recurrencec | 102 (53.7) |
| Deaths due to HCC | 24 (12.6) |
|
SALL4 |
p-value |
K19a |
p-value |
EpCAMa |
p-value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | Negative | Positive | ||||
| Frequency | 151 (79.5) | 39 (20.5) | 159 (84.1) | 30 (15.9) | 97 (51.3) | 92 (48.7) | |||
| Age (yr) | .858 | .694 | .020 | ||||||
| < 60 | 81 (53.6) | 20 (51.3) | 83 (52.2) | 17 (56.7) | 43 (44.3) | 57 (62.0) | |||
| ≥ 60 | 70 (46.4) | 19 (48.7) | 76 (47.8) | 13 (43.3) | 54 (55.7) | 35 (38.0) | |||
| Sex | 1.000 | .808 | .113 | ||||||
| Male | 32 (21.2) | 8 (20.5) | 33 (20.8) | 7 (23.3) | 16 (16.5) | 24 (26.1) | |||
| Female | 119 (78.8) | 31 (79.5) | 126 (79.2) | 23 (76.7) | 81 (83.5) | 68 (73.9) | |||
| Etiology | .319 | .829 | .010 | ||||||
| Hepatitis B virus–related | 40 (26.5) | 14 (35.9) | 45 (29.3) | 9 (30.0) | 36 (37.1) | 18 (19.6) | |||
| Non-hepatitis B virus–related | 111 (73.5) | 25 (64.1) | 114 (71.7) | 8 (70.0) | 61 (62.9) | 74 (80.3) | |||
| Serum α-fetoprotein (IU/mL) (n = 132) | 1.000 | .032 | .023 | ||||||
| < 1,000 | 76 (76) | 25 (78.1) | 86 (80.4) | 14 (58.3) | 54 (85.7) | 46 (79.3) | |||
| ≥ 1,000 | 24 (24) | 7 (21.9) | 21 (19.6) | 10 (41.7) | 9 (14.3) | 22 (20.7) | |||
| Multiplicity | .825 | .328 | .372 | ||||||
| Absent | 120 (79.5) | 32 (82.1) | 129 (81.1) | 22 (73.3) | 80 (82.5) | 71 (77.2) | |||
| Present | 31 (20.5) | 7 (17.9) | 30 (18.9) | 8 (26.7) | 17 (17.5) | 21 (22.8) | |||
| Tumor size (cm)b | .432 | .076 | .873 | ||||||
| < 5 | 105 (69.5) | 30 (76.9) | 118 (74.2) | 17 (56.7) | 70 (72.2) | 65 (70.7) | |||
| ≥ 5 | 16 (30.5) | 9 (23.1) | 41 (25.8) | 13 (43.3) | 27 (27.8) | 27 (29.3) | |||
| Pathologic T stage | 1.000 | .011 | .266 | ||||||
| pT1/pT2 | 122 (80.8) | 32 (82.1) | 134 (84.3) | 19 (63.3) | 82 (84.5) | 71 (77.2) | |||
| pT3a/pT3b/pT4 | 29 (19.2) | 7 (17.9) | 25 (15.7) | 11 (36.7) | 15 (15.5) | 21 (22.8) | |||
| Edmondson-Steiner grade | .410 | .648 | .019 | ||||||
| I/II | 36 (23.8) | 12 (30.8) | 42 (26.4) | 6 (20.0) | 32 (33) | 16 (17.4) | |||
| III/IV | 115 (76.2) | 27 (69.2) | 117 (73.6) | 24 (80.0) | 65 (67) | 76 (82.6) | |||
| Microvascular invasion | .581 | .685 | .011 | ||||||
| Absent | 94 (62.3) | 22 (56.4) | 98 (61.6) | 17 (56.7) | 68 (70.1) | 47 (51.1) | |||
| Present | 57 (37.7) | 17 (43.6) | 61 (38.4) | 13 (43.3) | 29 (29.9) | 45 (48.9) | |||
| Major vessel invasionc | .781 | .010 | .366 | ||||||
| Absent | 134 (88.7) | 34 (87.2) | 145 (91.2) | 22 (73.3) | 88 (90.7) | 79 (85.9) | |||
| Present | 17 (11.3) | 5 (12.8) | 14 (8.8) | 8 (26.7) | 9 (9.3) | 13 (14.1) | |||
Values are presented as number (%).
HCC, hepatocellular carcinomas; SALL4, Sal-like protein 4; EpCAM, epithelial cellular adhesion molecule.
an = 189 (tissue core loss in 1 case);
bSize of largest tumor in case of multiple tumors;
cMain or first order branches of portal vein and/or one or more of right, middle or left hepatic veins.
- 1. Lee JS, Heo J, Libbrecht L, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med 2006; 12: 410-6. ArticlePubMedPDF
- 2. Nault JC, De Reynies A, Villanueva A, et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology 2013; 145: 176-87. ArticlePubMed
- 3. Woo HG, Lee JH, Yoon JH, et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res 2010; 70: 3034-41. ArticlePubMedPMCPDF
- 4. Yamashita T, Forgues M, Wang W, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res 2008; 68: 1451-61. PubMed
- 5. Kim H, Choi GH, Na DC, et al. Human hepatocellular carcinomas with “Stemness”-related marker expression: keratin 19 expression and a poor prognosis. Hepatology 2011; 54: 1707-17. ArticlePubMed
- 6. Kim H, Park YN. Hepatocellular carcinomas expressing ‘stemness’-related markers: clinicopathological characteristics. Dig Dis 2014; 32: 778-85. ArticlePubMedPDF
- 7. Govaere O, Komuta M, Berkers J, et al. Keratin 19: a key role player in the invasion of human hepatocellular carcinomas. Gut 2014; 63: 674-85. ArticlePubMedPMC
- 8. Kim H, Yoo JE, Cho JY, et al. Telomere length, TERT and shelterin complex proteins in hepatocellular carcinomas expressing “stemness”-related markers. J Hepatol 2013; 59: 746-52. ArticlePubMed
- 9. Oikawa T, Kamiya A, Zeniya M, et al. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology 2013; 57: 1469-83. ArticlePubMed
- 10. Yong KJ, Gao C, Lim JS, et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med 2013; 368: 2266-76. ArticlePubMedPMC
- 11. Zeng SS, Yamashita T, Kondo M, et al. The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. J Hepatol 2014; 60: 127-34. ArticlePubMed
- 12. Zhang J, Tam WL, Tong GQ, et al. SALL4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol 2006; 8: 1114-23. ArticlePubMedPDF
- 13. Wu Q, Chen X, Zhang J, et al. SALL4 interacts with Nanog and cooccupies Nanog genomic sites in embryonic stem cells. J Biol Chem 2006; 281: 24090-4. ArticlePubMed
- 14. Kohlhase J, Heinrich M, Schubert L, et al. Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet 2002; 11: 2979-87. ArticlePubMed
- 15. Ikeda H, Sato Y, Yoneda N, et al. α-Fetoprotein-producing gastric carcinoma and combined hepatocellular and cholangiocarcinoma show similar morphology but different histogenesis with respect to SALL4 expression. Hum Pathol 2012; 43: 1955-63. ArticlePubMed
- 16. Ushiku T, Shinozaki A, Shibahara J, et al. SALL4 represents fetal gut differentiation of gastric cancer, and is diagnostically useful in distinguishing hepatoid gastric carcinoma from hepatocellular carcinoma. Am J Surg Pathol 2010; 34: 533-40. ArticlePubMed
- 17. Ma Y, Cui W, Yang J, et al. SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood 2006; 108: 2726-35. ArticlePubMedPMC
- 18. Cao D, Humphrey PA, Allan RW. SALL4 is a novel sensitive and specific marker for metastatic germ cell tumors, with particular utility in detection of metastatic yolk sac tumors. Cancer 2009; 115: 2640-51. ArticlePubMed
- 19. Oikawa T, Kamiya A, Kakinuma S, et al. SALL4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology 2009; 136: 1000-11. ArticlePubMed
- 20. Liu TC, Vachharajani N, Chapman WC, Brunt EM. SALL4 immunoreactivity predicts prognosis in Western hepatocellular carcinoma patients but is a rare event: a study of 236 cases. Am J Surg Pathol 2014; 38: 966-72. PubMedPMC
- 21. Gonzalez-Roibon N, Katz B, Chaux A, et al. Immunohistochemical expression of SALL4 in hepatocellular carcinoma, a potential pitfall in the differential diagnosis of yolk sac tumors. Hum Pathol 2013; 44: 1293-9. ArticlePubMed
REFERENCES
Figure & Data
References
Citations

- Cytoplasmic SALL4-A isoform expression as a diagnostic marker of less aggressive tumor behavior in gastric cancer
Saeed Rahmani, Amirhesam Babajani, Maryam Abolhasani, Roya Ghods, Elham Kalantari, Zahra Madjd
World Journal of Surgical Oncology.2025;[Epub] CrossRef - Stemness markers in hepatocellular carcinoma of Eastern vs. Western population: Etiology matters?
Caecilia HC Sukowati, Korri El-Khobar, Chyntia Olivia Maurine Jasirwan, Juferdy Kurniawan, Rino Alvani Gani
Annals of Hepatology.2024; 29(1): 101153. CrossRef - Research progress and prospects of AFP-positive gastric cancer
Long Zhao, Changjiang Yang, Yilin Lin, Shan Wang, Yingjiang Ye, Zhanlong Shen
Foregut Surgery.2022; 2(1): 29. CrossRef - SALL4 and microRNA: The Role of Let-7
Jun Liu, Madeline A. Sauer, Shaza G. Hussein, Junyu Yang, Daniel G. Tenen, Li Chai
Genes.2021; 12(9): 1301. CrossRef - Hepatoid Teratoma, Hepatoid Yolk Sac Tumor, and Hepatocellular Carcinoma
Khaleel I. Al-Obaidy, Sean R. Williamson, Nathan Shelman, Muhammad T. Idrees, Thomas M. Ulbright
American Journal of Surgical Pathology.2021; 45(1): 127. CrossRef - Targeting an Inducible SALL4-Mediated Cancer Vulnerability with Sequential Therapy
Junyu Yang, Chong Gao, Miao Liu, Yao-Chung Liu, Junsu Kwon, Jun Qi, Xi Tian, Alicia Stein, Yanjing V. Liu, Nikki R. Kong, Yue Wu, Shenyi Yin, Jianzhong Xi, Zhiyuan Chen, Kalpana Kumari, Hannan Wong, Hongbo Luo, Leslie E. Silberstein, Julie A.I. Thoms, Ash
Cancer Research.2021; 81(23): 6018. CrossRef - Lipoprotein‐Like Nanoparticle Carrying Small Interfering RNA Against Spalt‐Like Transcription Factor 4 Effectively Targets Hepatocellular Carcinoma Cells and Decreases Tumor Burden
William Cruz, Huang Huang, Brian Barber, Elisa Pasini, Lili Ding, Gang Zheng, Juan Chen, Mamatha Bhat
Hepatology Communications.2020; 4(5): 769. CrossRef - Hepatoid adenocarcinoma of the stomach: a unique subgroup with distinct clinicopathological and molecular features
Yakun Wang, Li Sun, Zhongwu Li, Jing Gao, Sai Ge, Cheng Zhang, Jiajia Yuan, Xicheng Wang, Jian Li, Zhihao Lu, Jifang Gong, Ming Lu, Jun Zhou, Zhi Peng, Lin Shen, Xiaotian Zhang
Gastric Cancer.2019; 22(6): 1183. CrossRef - Gynecologic Serous Carcinoma: An Immunohistochemical Analysis of Malignant Body Fluid Specimens
Shuyue Ren, William Klump
Archives of Pathology & Laboratory Medicine.2019; 143(6): 677. CrossRef - The Pluripotency Network in Colorectal Cancer Pathogenesis and Prognosis: an Update
Ioannis A Voutsadakis
Biomarkers in Medicine.2018; 12(6): 653. CrossRef - Cancer stem cells in hepatocellular carcinoma: an overview and promising therapeutic strategies
Nuozhou Wang, Shanshan Wang, Ming-Yue Li, Bao-guang Hu, Li-ping Liu, Sheng-li Yang, Shucai Yang, Zhongqin Gong, Paul B. S. Lai, George G. Chen
Therapeutic Advances in Medical Oncology.2018;[Epub] CrossRef - DNA demethylation induces SALL4 gene re-expression in subgroups of hepatocellular carcinoma associated with Hepatitis B or C virus infection
H Fan, Z Cui, H Zhang, S K Mani, A Diab, L Lefrancois, N Fares, P Merle, O Andrisani
Oncogene.2017; 36(17): 2435. CrossRef - Higher expression of SALL4 predicts poor cancer prognosis: A meta-analysis
Hongyu Shen, Liangpeng Li, Dandan Wang, Sujin Yang, Xiu Chen, Siying Zhou, Shanliang Zhong, Jianhua Zhao, Jinhai Tang
Cancer Biomarkers.2017; 19(4): 365. CrossRef - SALL4 suppresses PTEN expression to promote glioma cell proliferation via PI3K/AKT signaling pathway
Chuanjin Liu, Haibin Wu, Yanyan Li, Liang Shen, Renchun Yu, Hongwei Yin, Ting Sun, Chunming Sun, Youxin Zhou, Ziwei Du
Journal of Neuro-Oncology.2017; 135(2): 263. CrossRef - Liver Cancer: Molecular Characterization, Clonal Evolution and Cancer Stem Cells
Germana Castelli, Elvira Pelosi, Ugo Testa
Cancers.2017; 9(9): 127. CrossRef - Oncofetal gene SALL4 and prognosis in cancer: A systematic review with meta-analysis
Lorenzo Nicolè, Tiziana Sanavia, Nicola Veronese, Rocco Cappellesso, Claudio Luchini, Paolo Dabrilli, Ambrogio Fassina
Oncotarget.2017; 8(14): 22968. CrossRef - SALL4, the missing link between stem cells, development and cancer
Hiro Tatetsu, Nikki R. Kong, Gao Chong, Giovanni Amabile, Daniel G. Tenen, Li Chai
Gene.2016; 584(2): 111. CrossRef - A New Cell Block Method for Multiple Immunohistochemical Analysis of Circulating Tumor Cells in Patients with Liver Cancer
Soo Jeong Nam, Hyun Yang Yeo, Hee Jin Chang, Bo Hyun Kim, Eun Kyung Hong, Joong-Won Park
Cancer Research and Treatment.2016; 48(4): 1229. CrossRef - Functional and clinical significance of SALL4 in breast cancer
Ebubekir Dirican, Mustafa Akkiprik
Tumor Biology.2016; 37(9): 11701. CrossRef - MicroRNA-33b suppresses the proliferation and metastasis of hepatocellular carcinoma cells through the inhibition of Sal-like protein 4 expression
Qinggang Tian, Yao Xiao, Yanting Wu, Yun Liu, Zhiqing Song, Wenfeng Gao, Jing Zhang, Jingling Yang, Yuguo Zhang, Tuankui Guo, Furong Dai, Zhigang Sun
International Journal of Molecular Medicine.2016; 38(5): 1587. CrossRef - Oncogenic protein SALL4 and ZNF217 as prognostic indicators in solid cancers: a meta-analysis of individual studies
Ji Cheng, Jinbo Gao, Xiaoming Shuai, Kaixiong Tao
Oncotarget.2016; 7(17): 24314. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure




Fig. 1.
Fig. 2.
Fig. 3.
Fig. 4.
| Characteristic | No. (%) |
|---|---|
| Age (yr) | 58.1 ± 11.8 |
| Sex | |
| Male | 151 (79.5) |
| Female | 39 (20.1) |
| Etiology | |
| Hepatitis B | 136 (71.6) |
| Hepatitis C | 17 (8.9) |
| Alcohol | 8 (4.2) |
| Others | 30 (15.8) |
| Serum alpha-fetoprotein (IU/mL) | 1,634.6 ± 4,590.4 |
| Serum PIVKA-II (AU/mL) | 661.2 ± 1,528.9 |
| Multiplicity | |
| Absent | 106 (55.8) |
| Present | 31 (16.3) |
| Tumor size (cm) |
4.6 ± 3.0 |
| Edmondson-Steiner grade | |
| I | 1 (0.5) |
| II | 47 (24.7) |
| III | 122 (64.2) |
| IV | 20 (10.5) |
| Microvascular invasion | |
| Absent | 116 (61.1) |
| Present | 74 (38.9) |
| Major vessel invasion |
|
| Absent | 168 (88.4) |
| Present | 22 (11.6) |
| Follow-up | |
| Recurrence |
102 (53.7) |
| Deaths due to HCC | 24 (12.6) |
| p-value | Hazard ratio | 95% Confidence interval |
||
|---|---|---|---|---|
| Lower | Upper | |||
| Overall survival | ||||
| SALL4 positivity | .004 | 3.556 | 1.511 | 8.367 |
| Multiplicity of tumor | .016 | 2.929 | 1.223 | 7.014 |
| High T stage (T3 and 4) | .004 | 3.583 | 1.486 | 8.636 |
| Disease-free survival | ||||
| Size (> 3 cm) | .019 | 1.703 | 1.092 | 2.656 |
| Multiplicity of tumor | .004 | 1.937 | 1.234 | 3.039 |
| SALL4 |
p-value | K19 |
p-value | EpCAM |
p-value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | Negative | Positive | ||||
| Frequency | 151 (79.5) | 39 (20.5) | 159 (84.1) | 30 (15.9) | 97 (51.3) | 92 (48.7) | |||
| Age (yr) | .858 | .694 | .020 | ||||||
| < 60 | 81 (53.6) | 20 (51.3) | 83 (52.2) | 17 (56.7) | 43 (44.3) | 57 (62.0) | |||
| ≥ 60 | 70 (46.4) | 19 (48.7) | 76 (47.8) | 13 (43.3) | 54 (55.7) | 35 (38.0) | |||
| Sex | 1.000 | .808 | .113 | ||||||
| Male | 32 (21.2) | 8 (20.5) | 33 (20.8) | 7 (23.3) | 16 (16.5) | 24 (26.1) | |||
| Female | 119 (78.8) | 31 (79.5) | 126 (79.2) | 23 (76.7) | 81 (83.5) | 68 (73.9) | |||
| Etiology | .319 | .829 | .010 | ||||||
| Hepatitis B virus–related | 40 (26.5) | 14 (35.9) | 45 (29.3) | 9 (30.0) | 36 (37.1) | 18 (19.6) | |||
| Non-hepatitis B virus–related | 111 (73.5) | 25 (64.1) | 114 (71.7) | 8 (70.0) | 61 (62.9) | 74 (80.3) | |||
| Serum α-fetoprotein (IU/mL) (n = 132) | 1.000 | .032 | .023 | ||||||
| < 1,000 | 76 (76) | 25 (78.1) | 86 (80.4) | 14 (58.3) | 54 (85.7) | 46 (79.3) | |||
| ≥ 1,000 | 24 (24) | 7 (21.9) | 21 (19.6) | 10 (41.7) | 9 (14.3) | 22 (20.7) | |||
| Multiplicity | .825 | .328 | .372 | ||||||
| Absent | 120 (79.5) | 32 (82.1) | 129 (81.1) | 22 (73.3) | 80 (82.5) | 71 (77.2) | |||
| Present | 31 (20.5) | 7 (17.9) | 30 (18.9) | 8 (26.7) | 17 (17.5) | 21 (22.8) | |||
| Tumor size (cm) |
.432 | .076 | .873 | ||||||
| < 5 | 105 (69.5) | 30 (76.9) | 118 (74.2) | 17 (56.7) | 70 (72.2) | 65 (70.7) | |||
| ≥ 5 | 16 (30.5) | 9 (23.1) | 41 (25.8) | 13 (43.3) | 27 (27.8) | 27 (29.3) | |||
| Pathologic T stage | 1.000 | .011 | .266 | ||||||
| pT1/pT2 | 122 (80.8) | 32 (82.1) | 134 (84.3) | 19 (63.3) | 82 (84.5) | 71 (77.2) | |||
| pT3a/pT3b/pT4 | 29 (19.2) | 7 (17.9) | 25 (15.7) | 11 (36.7) | 15 (15.5) | 21 (22.8) | |||
| Edmondson-Steiner grade | .410 | .648 | .019 | ||||||
| I/II | 36 (23.8) | 12 (30.8) | 42 (26.4) | 6 (20.0) | 32 (33) | 16 (17.4) | |||
| III/IV | 115 (76.2) | 27 (69.2) | 117 (73.6) | 24 (80.0) | 65 (67) | 76 (82.6) | |||
| Microvascular invasion | .581 | .685 | .011 | ||||||
| Absent | 94 (62.3) | 22 (56.4) | 98 (61.6) | 17 (56.7) | 68 (70.1) | 47 (51.1) | |||
| Present | 57 (37.7) | 17 (43.6) | 61 (38.4) | 13 (43.3) | 29 (29.9) | 45 (48.9) | |||
| Major vessel invasion |
.781 | .010 | .366 | ||||||
| Absent | 134 (88.7) | 34 (87.2) | 145 (91.2) | 22 (73.3) | 88 (90.7) | 79 (85.9) | |||
| Present | 17 (11.3) | 5 (12.8) | 14 (8.8) | 8 (26.7) | 9 (9.3) | 13 (14.1) | |||
Values are presented as mean ± standard deviation or number (%). HCC, hepatocellular carcinomas. Size of largest tumor in case of multiple tumors; Main or first order branches of portal vein and/or one or more of right, middle or left hepatic veins; Local recurrence or distant metastasis.
SALL4, Sal-like protein 4.
Values are presented as number (%). HCC, hepatocellular carcinomas; SALL4, Sal-like protein 4; EpCAM, epithelial cellular adhesion molecule. n = 189 (tissue core loss in 1 case); Size of largest tumor in case of multiple tumors; Main or first order branches of portal vein and/or one or more of right, middle or left hepatic veins.

 E-submission
E-submission