Current status and future perspectives of liquid biopsy in non-small cell lung cancer
Article information
Abstract
With advances in target therapy, molecular analysis of tumors is routinely required for treatment decisions in patients with advanced non-small cell lung cancer (NSCLC). Liquid biopsy refers to the sampling and analysis of circulating cell-free tumor DNA (ctDNA) in various body fluids, primarily blood. Because the technique is minimally invasive, liquid biopsies are the future in cancer management. Epidermal growth factor receptor (EGFR) ctDNA tests have been performed in routine clinical practice in advanced NSCLC patients to guide tyrosine kinase inhibitor treatment. In the near future, liquid biopsy will be a crucial prognostic, predictive, and diagnostic method in NSCLC. Here we present the current status and future perspectives of liquid biopsy in NSCLC.
Molecular analysis is traditionally performed on tumor tissue. Although the number of mandatory tests for treatment decisions increases in patients with advanced non-small cell lung cancer (NSCLC), it is difficult to secure adequate tumor tissue for this purpose [1]. Small biopsy specimens, cell blocks, or aspirates are often the only available samples in patients with advanced NSCLC [2,3]. It is difficult to repeat tissue biopsies because they are invasive. Liquid biopsy could be an alternative or a complementary minimally invasive method for detecting molecular changes in NSCLC [1,2,4,5].
The clinical use of liquid biopsy to select patients with advanced NSCLC who are candidates for third-generation epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) therapy has been demonstrated in many clinical trials [6-10]. The United States Food and Drug Administration (FDA) approved Cobas EGFR Mutation Test v2 (Roche, Indianapolis, IN, USA) in 2018 as a companion diagnostic for third-generation EGFR TKI based on these results [11]. The Korea National Health Insurance Service (NHIS) has covered circulating cell-free tumor DNA (ctDNA) tests for EGFR mutations in advanced NSCLC since 2018. In this review, we present the current status and future perspectives of liquid biopsy in patients with NSCLC.
BIOLOGY OF CIRCULATING TUMOR DNA
Liquid biopsy refers to the collection and analysis of analytes from various body fluids such as blood, urine, sputum, and pleural fluid [12-14]. Different analytes can be present in a liquid biopsy including circulating tumor cells (CTCs), circulating cell-free DNAs (cfDNAs), circulating tumor RNAs (ctRNAs), circulating exosomes, tumor-educated platelets, proteins, and metabolites [15,16]. CTCs are intact, viable tumor cells circulating in the blood [12]. Cancer releases single or clusters of CTCs into the bloodstream during the course of hematogenous spread. cfDNA refers to all circulating DNA in body fluids. cfDNA can be derived from neoplastic as well as non-neoplastic cells [15,16]. cfDNA can be detected in other body fluids, including urine, saliva, or cerebrospinal fluid. ctDNA refers to a subgroup of cell-free DNA originating from tumor cells.
Circulating DNA fragments have a fragmentation pattern similar to a nucleosomal fragmentation pattern resulting from activation of nucleases in apoptotic cells [17,18]. Apoptosis (and necrosis) of the tumor is thought to be the major source of ctDNA [19-21]. As the tumor grows, apoptosis/necrosis increases as a result of rapid cell turnover. This leads to more release of tumor DNA into the circulation [14]. Macrophages may play a role in tumor cell release by phagocytosis of necrotic tumor cells. CTCs and active secretion from tumor cells may also be a source of ctDNA [21,22]. Circulating DNA is rapidly cleared via the kidney, liver, and spleen [23,24]. The load of ctDNA is highly correlated with total tumor burden and both volume and number of metastatic sites, suggesting that ctDNA may potentially have diagnostic and prognostic value [1,25-27]. Abbosh et al. [25] monitored clonal changes in NSCLC cells from initial diagnosis to death in the TRACER × trial (TRAcking non-small cell lung cancer evolution through therapy R[x]) and suggested that ctDNA release is dependent on proliferation rate, apoptotic potential, and genomic instability. The amount of ctDNA before treatment highly correlated with the metabolic tumor volume on positron emission tomography–computed tomography. However, some patients with metastases have an unexpectedly low fraction of ctDNA [17,28,29]. Further investigation is required to understand the release and removal of ctDNA.
LIQUID BIOPSY VERSUS TISSUE BIOPSY
Liquid biopsy is a minimally invasive procedure [30], it avoids the complications of surgical biopsies and can be used for serial monitoring. Liquid biopsy allows for storage of tissue for further analyses such as immunohistochemistry related to immuno-oncology or participation in clinical trials. Tumors generally consist of different subclones (tumor heterogeneity). The outgrowth of some subclones under selection pressures such as therapeutic stress, particularly by targeted drugs, and micro-environmental changes can lead to disease progression and metastasis [14,30,31]. This clonal evolution can dynamically modify the genomic landscape of tumors. Tissue-based molecular analysis provides only a snapshot of tumor heterogeneity when and where the tumor was biopsied. Liquid biopsy can analyze complete and real-time molecular profiling of the tumor because blood samples contain ctDNA constantly released into the circulation from multiple regions of primary and metastatic tumors [14].
Despite these advantages, liquid biopsy has several limitations in its widespread use. ctDNA detection requires more sensitive techniques than traditional approaches such as Sanger sequencing or pyrosequencing because of low fraction and high fragmentation of ctDNA [17,18,21,30]. Highly sensitive and highly specific tests, which are not available in all laboratories, are needed for ctDNA analysis. Liquid biopsy has an unfamiliar preanalytical variable associated with special processing and handling [11,32]. It does not yield information concerning histological tumor type, morphologic changes such as small cell lung cancer transformation that is one of the acquired resistance mechanisms to EGFR TKIs, and the tumor microenvironment in a liquid biopsy [16,33].
CURRENT TECHNOLOGIES FOR CIRCULATING TUMOR DNA DETECTION
Current tools for ctDNA analysis include real-time polymerase chain reaction (RT-PCR), digital PCR (dPCR), and next-generation sequencing (NGS) [15,21,30,34]. RT-PCR and dPCR are targeted methods that only allow screening of specific mutations. NGS allows targeted sequencing and whole exome sequencing. RT-PCR detects allele frequency (AF) and the ratio of variant alleles to wild type (WT) alleles down to 0.1% [8,15,35]. NGS and dPCR have detection thresholds approximately 0.01% or lower [36]. NGS and dPCR allow more precise quantification of the amount of ctDNA harboring target mutations than RT-PCR. dPCR can determine the absolute concentration of a gene of interest in a sample using the ratio of positive partitions with a targeted PCR product and an associated fluorescence signal over the total number [37]. RT-PCR uses a fluorescence readout to measure the amount of PCR product after each amplification and calculates the relative ratio of the target and reference genes for each sample (Semi-Quantitative Index).
The clinical use of various ctDNA tests for detecting EGFR mutations in plasma from patients with advanced NSCLC has been validated [6-9,38]. A meta-analysis showed that the sensitivity of ctDNA testing for the EGFR mutation was 66.4% (95% confidence interval [CI], 62.7% to 69.9%) and specificity was 95.6% (95% CI, 83.3% to 99.0%) in lung adenocarcinoma [39].
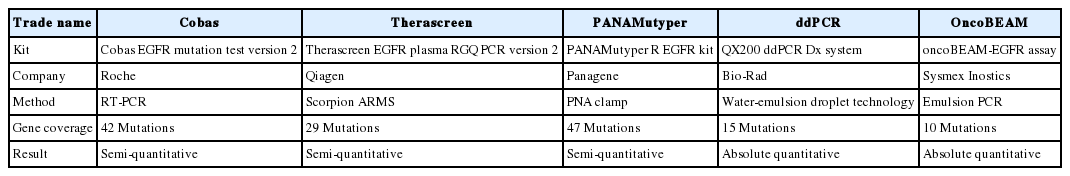
The Cobas EGFR Mutation Test v2 is the first ctDNA test to be approved by the US FDA to select patients who may benefit from TKI treatment. The Cobas EGFR Mutation Test v2 (Cobas) and PANAMutyper R EGFR (PANAMutyper; Panagene, Daejeon, Korea) were approved by the Korea FDA. A cross-platform comparison study showed comparable sensitivity between Cobas and PANAMutyper [40]. Commercially available ctDNA tests for detecting EGFR mutations in plasma are summarized in Table 1 [11,14,40-42].
Cobas can detect 42 mutations in exons 18–21 including L858R, exon 19 deletions, L861Q, and T790M in tissue and plasma [40]. The Cobas DNA Sample Preparation Kit (Roche, Indianapolis, IN, USA) was simultaneously developed for extracting ctDNA from plasma. PANAMutyper is a combination of peptide nucleic acid (PNA)-mediated clamping and melting curve analysis [40]. PNAClamp is a technology that selectively amplifies only the desired mutant sequences by combining the PNA clamping probe complementary to the WT sequences [40]. The PNA probe has a unique melting temperature according to the base sequence, separating from the DNA of the target sequence at a given temperature and reducing the fluorescence signal. The genotype of the target DNA can be determined by analyzing the temperature at which the signal decreases. The Therascreen EGFR Plasma RGQ PCR Kit (Qiagen, Manchester, UK) can detect EGFR mutations with amplification-refractory mutation system PCR and scorpion primers.
Droplet digital PCR (ddPCR; Bio-Rad, Hercules, CA, USA) and OncoBEAM (beads, emulsion, amplification, and magnetics; Sysmex Inostics, Hamburg, Germany) are dPCR-based techniques.
In dPCR, the sample is distributed into many reaction compartments containing one or no DNA copies and associated PCR reagents [4,12,40]. After amplification, the individual compartments are analyzed by a binary (presence or absence of amplification product) system. The number of compartments with amplification product corresponds directly to the number of copies of target mutation in the sample. In ddPCR, emulsion droplets are analyzed by fluorescence signal detection after target DNA is amplified within water-in-oil emulsion droplets. OncoBEAM combines emulsion PCR with magnetic beads and flow cytometry. After amplification, beads are coated with thousands of copies of single DNA molecules and each bead enters the water droplets [43]. Flow cytometry analyzes the beads. A prospective study revealed that the positive predictive value is 100% for detection of EGFR L858R and 19del and 79% for T790M [27].
Because there is an increasing number of mutations to be analyzed, NGS panels are an attractive approach for ctDNA analysis [15]. NGS panels offer advantages of higher throughput, higher sensitivity, more efficient use of limited tissue, and lower cost per analysis than PCR-based methods [15,44,45]. Foundation-One CDx (Foundation Medicine, Cambridge, MA, USA) is an FDA-approved targeted NGS panel and detects mutations in 324 genes. Another FDA-approved panel, OncomineDx Target Test (ThermoFisher Scientific, Waltham, MA, USA) is designed to detect 368 variants in 23 genes associated with NSCLC including EGFR, ROS1, and BRAF. The overall concordance rate of gene panel testing for tissue and ctDNA was 70.3% for EGFR mutation [46]. Cancer personalized profiling by deep sequencing combines optimized library preparation methods for low DNA input with a specialized bioinformatics approach to design a “selector” consisting of biotinylated DNA oligonucleotides that target recurrently mutated genomic regions in the cancer of interest [26,47].
CURRENT USE OF CIRCULATING TUMOR DNA TESTING IN LUNG CANCER
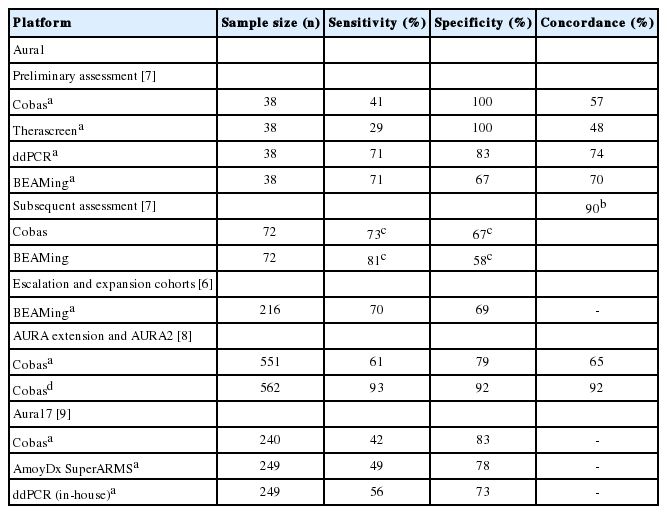
The clinical use of ctDNA testing for EGFR mutation using plasma specimens has been demonstrated by the AURA trials, which evaluated the dose, safety, and efficacy of osimertinib in patients with NSCLC who progressed following EGFR TKIs. The sensitivity of Cobas, OncoBEAM, ddPCR, and Therascreen for plasma T790M was up to 93%, 81%, 71%, and 29%, respectively (Table 2) [6-10]. The specificity of Cobas, OncoBEAM, ddPCR, and Therascreen for plasma T790M was 100%, 69%, 83%, and 100%, respectively. The concordance rate of T790M testing in plasma and tissue was up to 74%. Patients with T790M mutation in tissues or plasma had similar outcomes of osimertinib treatment [6,7,10]. Aura 17 showed that outcomes for patients with T790M mutations were comparable across the three plasma tests (56%–64%) [10]. Patients with T790M-positive plasma/T790M-negative tissue had unfavorable outcomes [6]. Patients with T790M-positive tissue/T790M-negative plasma status had better outcomes than patients who were tissue- and plasmapositive [8].
Currently, plasma ctDNA tests are recommended to detect EGFR mutations in patients with advanced NSCLC [11,15,16,39].
Korea NHIS covers up to three uses of ctDNA testing to select NSCLC patients for EGFR TKI when a tissue biopsy specimen is unavailable or insufficient for molecular testing. Liquid samples other than plasma have not been approved. Cobas and PANAMutyper are ctDNA tests for EGFR mutation covered by Korea NHIS.
CURRENT STATUS OF LIQUID BIOPSY IN KOREA BASED ON EXTERNAL QUALITY ASSURANCE RESULTS
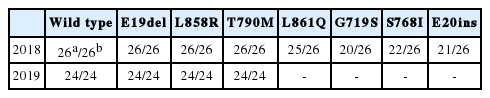
Proficiency testing for EGFR mutation analysis using liquid biopsy specimens has been performed annually by The Korean Society for Pathologists since 2018. Twenty-six laboratories participated in the proficiency test in 2018. The cfDNA Reference Standard (Horizon Discovery, Cambridge, UK) was used as reference material, which contained mutant AFs of 1% and 0% T790M, E19del, L858R, L861Q, G719S, S768I, and E20ins. Laboratories were requested to report the following: (1) time to sample processing, (2) tube type, (3) methods for plasma storage, (4) methods for ctDNA extraction, (5) methods for ctDNA analysis, and (6) results. All participating laboratories showed excellent performance with 100% accuracy for EGFR E19del, L858R, and T790M mutations (Table 3). There were no falsepositive results for L861Q, G719S, S768I, or E20ins. In 2019, 24 laboratories participated in the proficiency test. The Korea Research Institute of Standards and Science produced reference materials comprising AFs of 1.9% for E19del, 1.5% for L858R, and 2.5% for T790M. All participating laboratories had excellent performance with 100% accuracy for EGFR E19del, L858R, and T790M mutations (Table 3). The 2017 European External Quality Assessments (EQA) scheme for ctDNA analysis that involved 32 laboratories from 16 countries showed an overall error rate of 42% for 1% of T790M/L858R and 7% for 5% of T790M/L858R [48]. One false-positive result was observed. Compared with these results, the results of Korean EQA over the past 2 years support the notion that good quality ctDNA analyses for EGFR mutation are performed in Korean pathology laboratories.
PANAMutyper was the most commonly used test in 2018. Twenty-four of 26 laboratories (92%) used PANAMutyper for ctDNA analysis. One used Cobas and another used ddPCR. For ctDNA extraction, 12 of 26 laboratories (45%) used Cobas, nine (33%) used TANBead (Taiwan Advanced Nanotech, Taoyuan, Taiwan), three (11%) used Maxwell RSC ccfDNA Plasma Kit (Promega, Madison, WI, USA), 2 (7%) used QIAamp circulating nucleic acid kit (Qiagen), and one (4%) used LIBEX (Tianlong Science and Technology, Xian, China). Plasma volume used for ctDNA extraction was 2 mL in 17 (63%), 1 mL in 4 (15%), and 0.6 mL in six of the laboratories (22%). The volume of DNA used for mutation testing was 100 μL (n = 11, 41%) or 50 μL (n = 11, 41%). The purity of DNA was assessed by Nano-Drop (ThermoFisher Scientific) in 16 (76%) of the laboratories. Quibit (Invitrogen, Carlsbad, CA, USA), Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), Quantus (Promega), and TapeStation (Agilent Technologies) were also used in two, one, one, and one laboratories, respectively. Ethylenediaminetetraacetic acid (EDTA) tubes were used in 21 laboratories (78%). Seventeen laboratories (63%) kept blood samples refrigerated. Samples were processed within 2 hours in 13 (48%) and between 2 and 4 hours in 11 of the laboratories (41%). Plasma was stored in a freezer in 22 of the laboratories (82%) and immediately used in three laboratories.
Compared with 2018, there were decreases in sample processing time, decreases in volumes of plasma and DNA used, and increases in the use of TANBead for ctDNA extraction in 2019. All but one lab used PANAMutyper for ctDNA analysis. Twelve of 24 laboratories (50%) used TANBead for ctDNA extraction. The use of Cobas decreased (n = 6, 25%). Maxwell (n = 3, 13%), QIAamp (n = 1, 4%), and LIBEX (n = 2, 13%) were used. The number of laboratories using 2 mL of plasma for ctDNA extraction decreased to nine (38%), and those using 1 mL and 0.6 mL increased to seven (29%) and eight (33%), respectively. Some laboratories reduced the volume of DNA used for mutation testing to 50 μL (n = 18, 75%). Five laboratories (21%) used 100 μL. There was no change in DNA QC method. EDTA tubes were used in 21 laboratories (96%). Blood samples were kept refrigerated in 17 laboratories (71%). Twenty-one of the laboratories (88%) processed samples within 2 hours. The rest of the laboratories processed samples within 4 hours. The number of laboratories that immediately used plasma samples increased to six (25%). The rest stored plasma samples in a freezer. In the European EQA scheme, NGS (39%) is the most commonly used ctDNA test, followed by Cobas (26%) and ddPCR (23%) [48]. Fifty-five percent of the laboratories used QIAamp and 25% used Cobas for ctDNA extraction.
FUTURE PERSPECTIVES ON CIRCULATING TUMOR DNA TESTING IN LUNG CANCER
Other targetable mutations including ALK, BRAF, ROS1, MEK, and HER2 have been detected in plasma from patients with NSCLC, although the sensitivity for detecting these mutations is lower than that for EGFR mutant detection [15,49-52]. The NGS-based approaches will facilitate detection of various rare genetic mutations that could be targeted. Highly fragmented ctDNA could result in insufficient mappable sequences to identify fusion events [51]. New technologies have been developed for detection of ALK fusion in ctDNA or ctRNA [15,49-52]. Recently developed, hybrid capture-based NGS can retrieve large genomic fragments to whole genomes with high sequencing coverage and accurate detection of genomic alterations, including genomic re-arrangements and short variants at low AFs and copy number amplifications. The clinical value of rare targetable mutations has not been demonstrated in NSCLC. The clinical use of ctDNA testing for BRAF V600E mutation was reported in metastatic melanoma and similar clinical trials are in progress in NSCLC [53].
Sputum, pleural fluid, and urine could also be used for molecular analysis [15,39]. T790M mutation was detected in urine and relevant tumor tissue from patients with NSCLC undergoing TKI therapy [24]. Using extracellular vesicle-derived DNA and ctDNA from pleural fluid supernatants yielded significant improvement in EGFR mutation analysis compared to the use of cell blocks or smears [54]. Kawahara et al. [55] also detected EGFR mutations in four of 18 cytologically negative groups using pleural fluid supernatant ctDNA. Further trials using these body fluids are needed for use in real practice.
In the near future, ctDNA testing is expected to play a significant role in identifying prognostic, predictive, and diagnostic biomarkers in patients with NSCLC patients. Tracking of ctDNA levels by an absolute quantitative method could be a minimally invasive tool for monitoring therapeutic effects and tumor recurrence [2,15,16]. Clinical trials showed that serial ctDNA tests using BEAMing can be used to monitor tumor response during treatment. Median progression-free survival (PFS) was shortened and objective response rate decreased in patients with persistent detection of plasma T790M for six weeks after initiation of osimertinib treatment [56]. Changes in plasma EGFR mutation load predicted response in 93% and progression in 89% of patients with lung adenocarcinoma in advance of radiologic evaluation [57].
PFS was significantly longer for patients without ctDNA detection compared to those with ctDNA detection (295 vs. 55 days) during treatment [58]. However, the lack of consensus evaluation criteria and standard methodology are major limitations to expanding the use of ctDNA to monitor treatment efficacy and relapse [2].
Immunohistochemical programmed death-ligand 1 (PD-L1) assays are a companion or complementary diagnostic test for checkpoint inhibitor immune therapy. But, there are many challenges to the effective use of the PD-L1 test [59]. Tumor mutation burden (TMB), which is the total number of non-synonymous somatic mutations in the tumor genome, has recently been proposed as a predictive biomarker for immunotherapy in various tumors [58]. Liquid biopsy using the NGS-based approach is an emerging tool to assess TMB [2]. The OAK and POPLAR trials, which evaluated the efficacy of atezolizumab, showed a relationship between PFS and TMB of plasma ctDNA with FoundationOne CDx [60,61]. Giroux Leprieur et al. [62] also showed that TMB of ctDNA determined by an NCS gene panel approach can predict the efficacy of nivolumab in patients with advanced NSCLC. Patients with a ctDNA increase > 9% from baseline to the first tumor evaluation at 2 months had no long-term clinical benefit with nivolumab with a sensitivity of 71.4% and specificity of 100%. Because tumor-induced leukocyte infiltration may increase tumor volume or result in the development of new lesions, it may be difficult to radiographically assess the response to immunotherapy. Serial assessment of TMB using ctDNA testing will be beneficial for monitoring efficacy of immunotherapy. Goldberg et al. [63] showed that > 50% decrease in ctDNA was associated with improved PFS and OS. Median time to initial ctDNA response was 24.5 days, while the median time to initial radiographic partial response was 72.5 days. The use of ctDNA with imaging will allow a more comprehensive assessment of the response to immunotherapy.
The use of liquid biopsy has been studied in the assessment of minimal residual disease (MRD) after surgical resection in localized NSCLC [2,15]. Persistent detection of ctDNA after surgery in patients with NSCLC is highly correlated with tumor relapse [5,25,64]. Abbosh et al. [25] showed that the median interval between postoperative detection of ctDNA detection and tumor relapse confirmed on computed tomography (CT) imaging was 70 days in NSCLC patients. Chaudhuri et al. [64] also showed the utility of postoperative ctDNA detection using an NGS-based approach for MRD detection. The MRD detection rate was 94% by tracking all known mutations while 58% on average by tracking a single mutation. Postoperative ctDNA detection was earlier than imaging in 72% of patients with a median of 5.2 months. Patients without postoperative ctDNA detection had a better outcome than those with ctDNA detection, while radiographic response assessment by CT at four months was not prognostic. Prospective clinical trials are needed to establish the clinical utility of MRD assessment using ctDNA.
Low-dose CT scans play a significant role in reducing lung cancer mortality. However, the overall false-positive rate is 96.4% because of many indeterminate pulmonary nodules [65]. Conventional sputum cytology is a simple, rapid, and specific screening tool, but less sensitive for lung cancer. Epigenetic changes such as hypermethylation were detected in the sputum of patients with lung cancer years preceding clinical diagnosis [1,66,67]. Combining the detection of epigenetic changes in sputum and CT scanning could facilitate a reduction in false-positive results and improve early diagnosis of lung cancer. Low sensitivity is the major limitation for detection of hypermethylation because a relative excess of background WT DNAs can mask epigenetic changes. Panels of methylated promoter genes and ultrasensitive detection methods should be developed for lung cancer screening.
CONCLUSION
Although the mechanism of ctDNA release and removal is not clearly understood, EGFR mutation detection in liquid biopsy specimens can be used to guide EGFR TKI treatment when a tissue biopsy cannot be obtained. Liquid biopsy will play a significant role as a prognostic, predictive, and diagnostic tool for various tumors. Pathologists should be able to integrate the results of molecular tests from the liquid biopsy and the morphological characteristics of relevant tissue, understand the preanalytical variables and different assay performance, and participate in a multidisciplinary team approach for optimal management of cancer patients.
Notes
Author contributions
Conceptualization: SC, WSK, CHL.
Data curation: SC, JYH, YLC, WSK.
Formal analysis: SC, JYH, YLC, WSK.
Investigation: SC, JYH, YLC, WSK.
Methodology: YLC, WSK.
Project administration: WSK, CHL.
Supervision: WSK, CHL.
Writing—original draft: SC.
Writing—review & editing: SC, WSK.
Conflicts of Interest
The authors declare that they have no potential conflicts of interest.
Funding
No funding to declare.