Automated immunohistochemical assessment ability to evaluate estrogen and progesterone receptor status compared with quantitative reverse transcription-polymerase chain reaction in breast carcinoma patients
Article information
Abstract
Background
This study aimed to investigate the capability of an automated immunohistochemical (IHC) evaluation of hormonal receptor status in breast cancer patients compared to a well-validated quantitative reverse transcription–polymerase chain reaction (RT-qPCR) method.
Methods
This study included 93 invasive breast carcinoma cases that had both standard IHC assay and Oncotype Dx assay results. The same paraffin blocks on which Oncotype Dx assay had been performed were selected. Estrogen receptor (ER) and progesterone receptor (PR) receptor status were evaluated through IHC stains using SP1 monoclonal antibody for ER, and 1E2 monoclonal antibody for PR. All ER and PR immunostained slides were scanned, and invasive tumor areas were marked. Using the Quant-Center image analyzer provided by 3DHISTECH, IHC staining of hormone receptors was measured and converted to histochemical scores (H scores). Pearson correlation coefficients were calculated between Oncotype Dx hormone receptor scores and H scores, and between Oncotype Dx scores and Allred scores.
Results
H scores measured by an automated imaging system showed high concordance with RT-qPCR scores. ER concordance was 98.9% (92/93), and PR concordance was 91.4% (85/93). The correlation magnitude between automated H scores and RT-qPCR scores was high and comparable to those of Allred scores (for ER, 0.51 vs. 0.37 [p = .121], for PR, 0.70 vs. 0.72 [p = .39]).
Conclusions
Automated H scores showed a high concordance with quantitative mRNA expression levels measured by RT-qPCR.
Multimodality therapy has resulted in improved survival rates for breast cancer patients. Hormonal receptor status is especially important when considering therapeutic options and categorizing prognostically significant molecular subgroups. Routinely conducted immunohistochemistry plays a role in determining whether a patient needs anti-hormone therapy or not by measuring the expression of protein levels. To measure hormonal receptor status, a few scoring systems have been used, including the Allred score, histochemical scores (H scores), and quick score. The Allred and quick scores are semi-quantitative scores based on the sum of the percentage (PS) and intensity scores (IS). The Allred scoring system is a well-known, successfully clinically-validated scoring system [1]. An Allred score above 2, which corresponds to a weak staining intensity of greater than 1% of tumor cells, is the best cutoff for both disease-free survival and overall survival [2].
It is well established that multigene panels can accurately predict disease recurrence. Among them, Oncotype Dx has been widely used to determine high-risk groups for chemotherapy treatment since it was introduced [3]. The Oncotype Dx Recurrence Score (RS) is derived from quantitative measurement of mRNA expression that includes estrogen receptor (ER) and progesterone receptor (PR) and uses the quantitative reverse transcription–polymerase chain reaction (RT-qPCR) method. RS can predict anti-hormone therapy sensitivity in patients with ER-positive, node-negative breast cancer [4]. Previous studies have shown a high correlation between immunohistochemical scores and Oncotype Dx receptor scores [5–11]. Low levels of ER and PR are associated with high RS.
ER status is used as a dichotomous rather than a continuous variable when assessing patient suitability for anti-hormone therapy, and the degree of ER positivity has no impact on recommendations for the use of anti-hormonal therapy [12,13]. In a study conducted by Qureshi and Pervez [13], most tumors were either unequivocally ER-positive or ER-negative while weakly ER-positive tumors were rare [13]. Badve et al. [6] also stated that ER and PR by central immunohistochemical (IHC) were bimodal. However, some authors stated that ER expression is not bimodal in breast cancer [14].
The degree of nuclear expression measured by semi-quantitative scoring systems is dichotomous and skewed to a high score. In contrast, RT-qPCR methods can provide linear quantitative mRNA expression values that enable more precise decisions for clinicians and patients. However, not all patients can afford the high cost of these methods. If quantitative IHC scores show a good correlation with RT-qPCR results, they would accurately predict hormone receptor status and response to anti-hormone therapy.
The H score (histochemical score) is calculated by the sum of the proportion of tumor cells multiplied by the staining reactivity [1]. The score ranges from 0 to 300. A score of < 50 is considered negative and scores of 50–100, 101–200, and 201–300 are considered weakly positive (1+), moderately positive (2+), and strongly positive (3+), respectively [15].
We obtained the H score using a computer-aided image analysis program to secure faster and reproducible results. Computational approaches can play a role in better quantitative characterization of diseases and quantitative histomorphometry [16]. Current American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) recommendations encourage the use of quantitative image analysis techniques to improve the consistency of clinical interpretation [17]. ER and PR status assessment by image analysis presented an excellent agreement with visual histoscores and were predictive of recurrence-free survival and cancer-specific survival [18].
In our study, we compared the hormone scores of Oncotype Dx and the results of immunohistochemical expression scores—Allred score and computer-aided H score—and tested their agreements.
MATERIALS AND METHODS
Patient selection and data collection
Among those who had undergone surgery due to invasive breast carcinoma from 2014 to 2019 at Korea University Guro Hospital, 98-patient cases who had Oncotypes Dx test (Oncotype DX, Genomic Health, CA, USA) results were included. Five cases that were missing paraffin blocks were excluded. Eighty of the remaining 93 cases for which immunohistochemistry had been performed in the biopsy sample alone, were stained again in the paraffin block where Oncotype Dx was implemented.
Information such as patient age at diagnosis, tumor size, tumor grade, Ki-67 labeling index, and mitotic count was collected from pathologic review. ER score, PR score, and RS score data were collected from the Oncotype Dx report. To improve comparability, we stained the same paraffin block where the Oncotype Dx assay had been implemented. We also analyzed the whole invasive tumor area of the same section by obtaining the Allred score and computer-aided H score.
The clinical and pathologic characteristics of the final 93 cases are summarized in Table 1.
Immunohistochemical stain
The same paraffin blocks on which the Oncotype Dx assay was performed were selected. ER and PR receptor status were evaluated through immunohistochemical stains using the SP1 monoclonal antibody for ER, and the 1E2 monoclonal antibody for PR (Ventana Medical Systems, Tucson, AZ, USA). Formalin-fixed paraffin-embedded tissue samples were sliced with a microtome at 4 μm and placed on slides. The slides containing tissue sections were deparaffinized at 75°C, and cell conditioning was done with EDTA solution at 100°C for 4 minutes. Primary antibodies were applied for 20 minutes. A Ventana Benchmark Ultra instrument automatically stained the slides.
Allred score
The stained slides were reviewed and Allred scores for ER and PR were given by two skilled pathologists. Allred score was derived from the sum of PS (range, 0 to 5) and IS (range, 0 to 3).
Slide scanning and calculating IHC scores by image analysis
All ER and PR immunostained slides were scanned, and whole invasive tumor areas were marked by a pathologist. Using a QuantCenter image analyzer provided by 3DHISTECH (Budapest, Hungary), the results of the immunohistochemical staining of hormone receptors were measured and converted to H scores. The image analyzing system also provided automatically calculated Allred score results.
We set “score intensity” cutoff values in the QuantCenter program at 200, 160, and 100 to define negative, weakly positive, moderately positive, and strong positive staining intensity that corresponded to the reactivity of staining (0, 1, 2, 3, respectively) (255–200, 0; 200–160, 1; 160–100, 2; 100–0, 3).
Statistics
Pearson correlation coefficients were calculated between Oncotype Dx hormone receptor scores and H scores, and between Oncotype Dx scores and Allred scores. The RS score was also compared with IHC scores. Further, the automatically calculated Allred scores were compared with RT-qPCR scores and RS scores as well. Fisher’s z transformation was used to compare each correlation coefficient. Statistical analyses were performed with Graph-Pad Prism ver. 8.3 software (GraphPad Software Inc., San Diego, CA, USA).
The patients were subcategorized into a high score group (≥ 200), an intermediate score group (≥ 100 and < 200), and a low score group (< 100) to identify which subgroup was more correlated with the RT-qPCR score. The high score group (≥ 200) was subcategorized into < 250 and ≥ 250 groups.
RESULTS
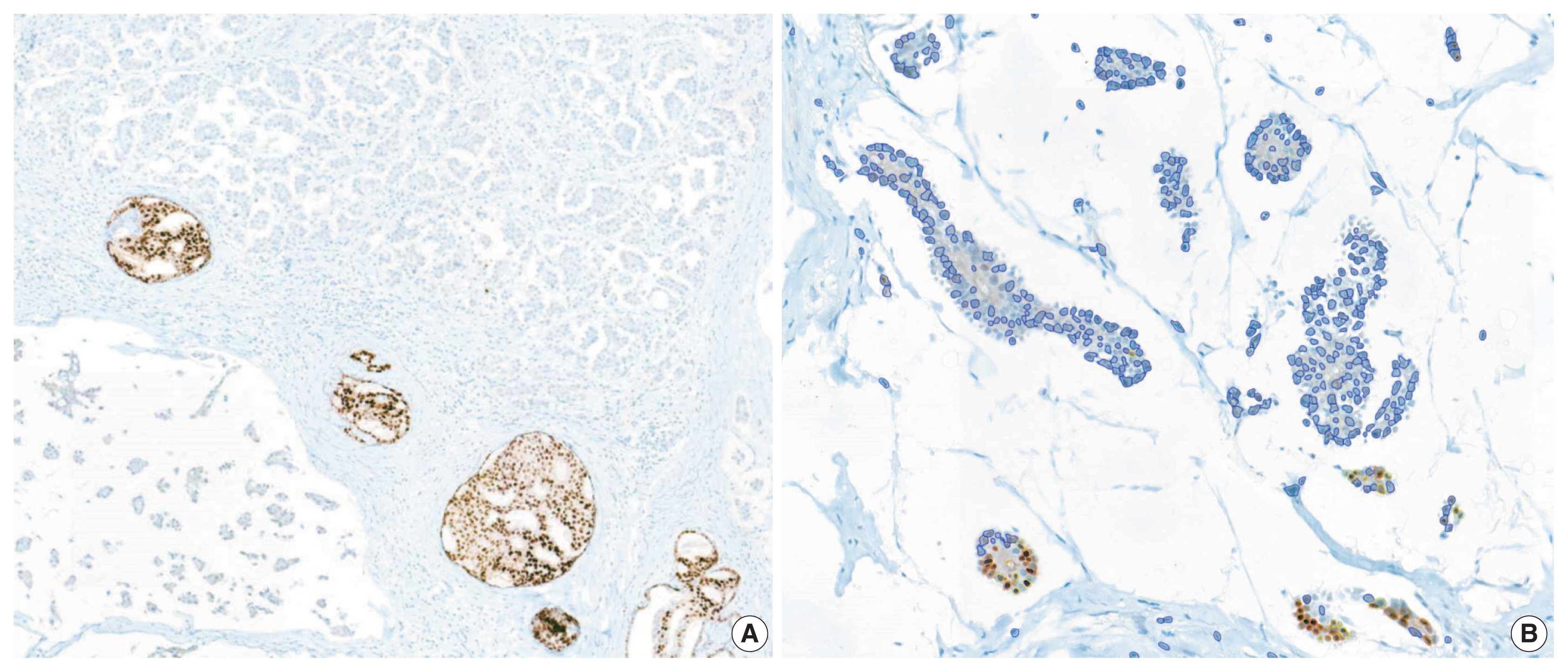
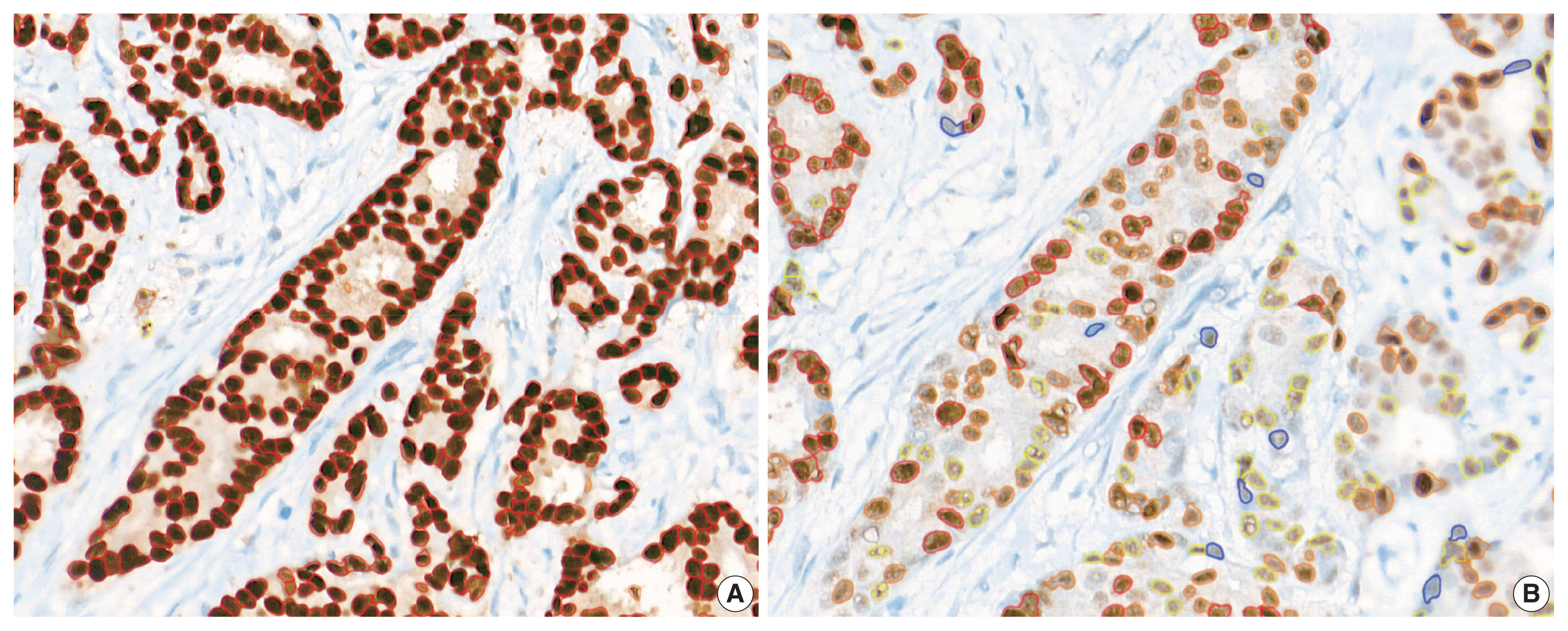
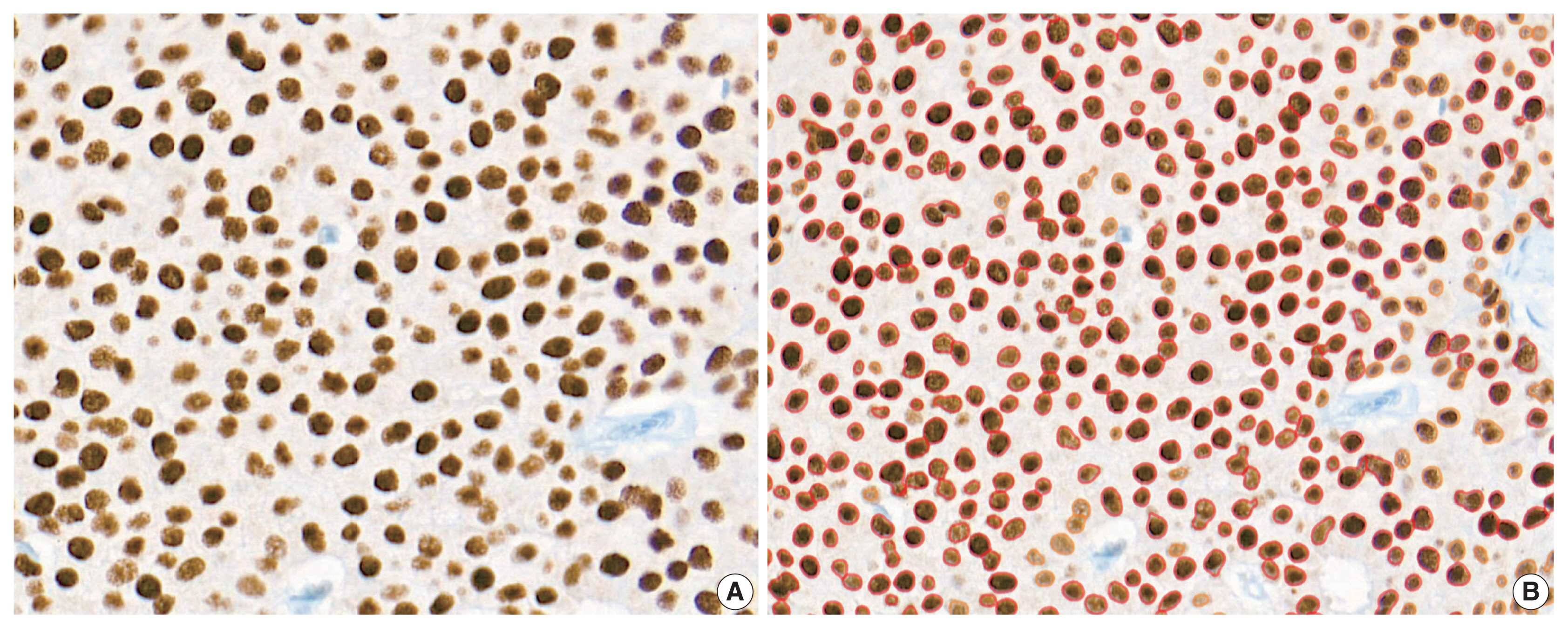
Immunohistochemical and RT-qPCR score results for ER and PR are summarized in Table 2. ER and PR concordance rate between the H score and the RT-qPCR assay was 98.9% (92/93) and 91.4% (85/93), respectively. The correlation coefficient between ER H score and ER RT-qPCR score was 0.51, and that between ER Allred score and ER RT-qPCR score was 0.37 (Table 3). The correlation coefficient between PR H score and PR RT-qPCR score was 0.70, and that between the PR Allred score and PR RT-qPCR score was 0.72. The correlation coefficients were higher for PR compared to ER (0.70 vs. 0.51 [p = .021] and 0.72 vs. 0.37 [p < .01]). The correlation coefficients for automatically calculated Allred scores were similar to those for the manual Allred score (Table 3). Fig. 1 demonstrates the correlation status between scores. Among all three measuring methods, the RT-qPCR score was closest to the normal distribution (Fig. 2). In general, the PR IHC stain showed a more heterogeneous staining intensity compared to ER IHC (Fig. 3). When we examined the cases of Allred score 8, computer-aided H score results showed a significant portion of moderately positive (intensity 2) nuclei as well as strong positive nuclei (intensity 3) (Fig. 4).
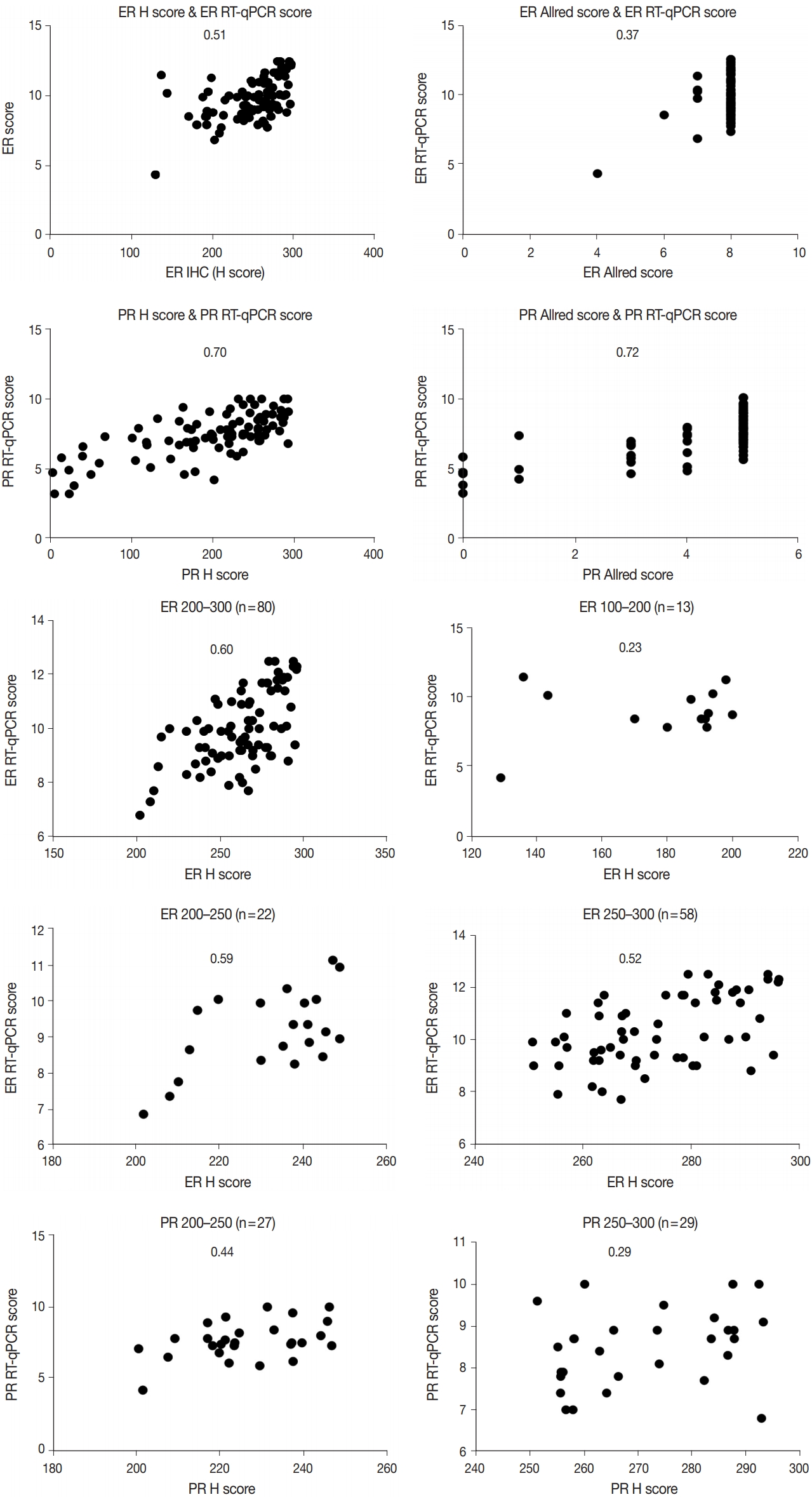
Correlation between IHC scores and RT-qPCR scores. The number in each graph indicates correlation coefficient (R). ER, estrogen receptor; PR, progesterone receptor; H, histochemical; RT-qPCR, quantitative reverse transcription-polymerase chain reaction; IHC, immunohistochemical.
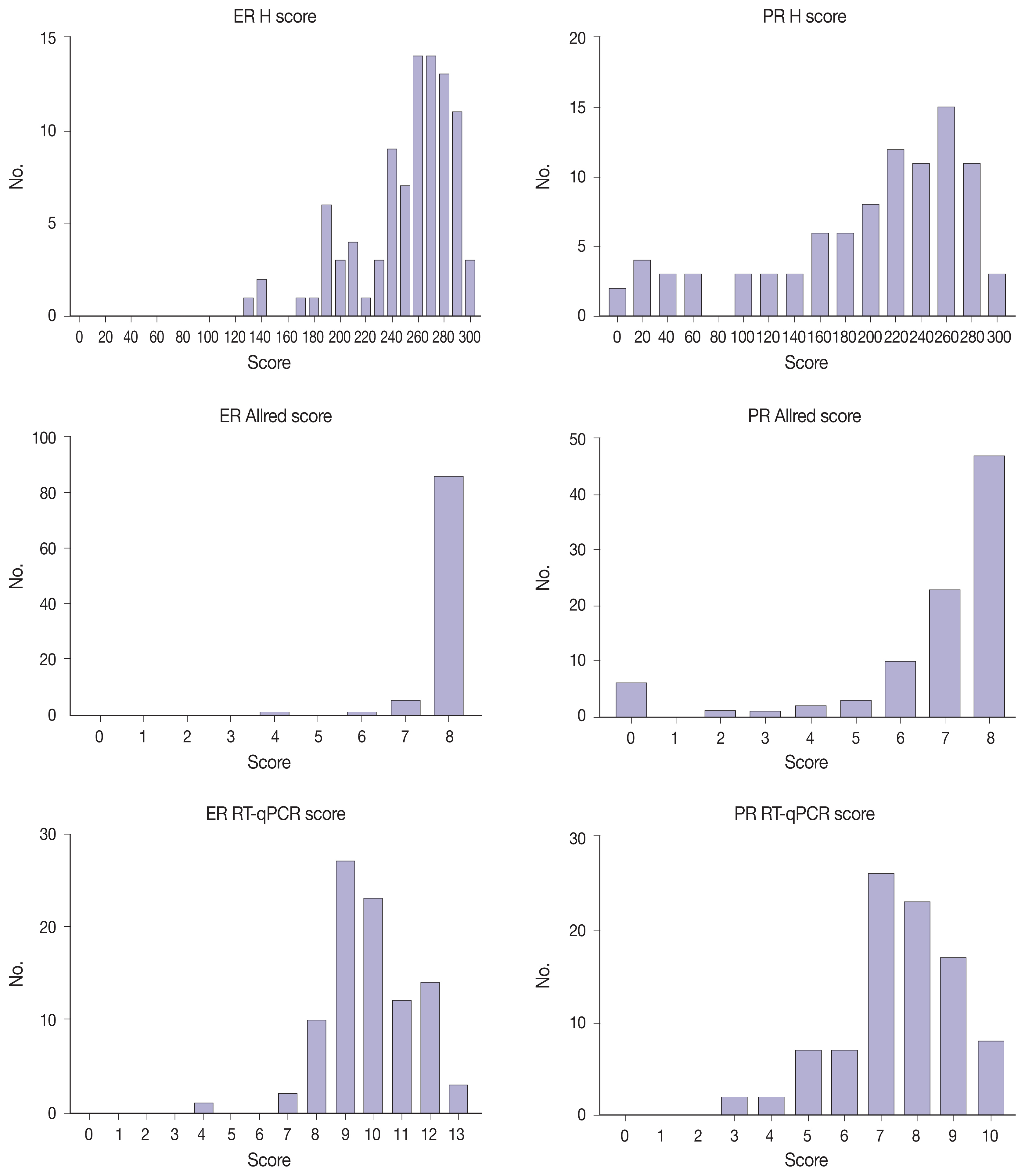
Distribution of H scores, Allred scores, and RT-qPCR scores. ER, estrogen receptor; PR, progesterone receptor; H, histochemical; RT-qPCR, quantitative reverse transcription-polymerase chain reaction; IHC, immunohistochemical.
PR-IHC stain showed more heterogeneous staining tendency (B) than ER-IHC stain (A) in the same tumor section. ER, estrogen receptor; PR, progesterone receptor; IHC, immunohistochemical.
(A) ER-stained slide with Allred score 8. (B) However, a significant portion of moderate nuclear staining (2+) was present as well as strong nuclear staining (3+) by image analysis. ER, estrogen receptor.
We inspected correlation magnitudes for each subgroup categorized by H score values. The intermediate H score group (range, 100 to 200) and low H score group (< 100) demonstrated the lowest correlation (Table 4). As the high H score group (range, 200 to 300) comprised a significant portion of all subjects, this group was further subcategorized into a 200–250 group and a 250–300 group. Compared to the 250–300 group, the 200–250 group showed a higher correlation with both ER and PR, although not statistically significant (0.59 vs. 0.52 for ER; 0.44 vs. 0.29 for PR) (p = .35 and p = .27) (Table 4).
There were eight discordant cases for PR, while there was one discordant case for ER (Table 5). For ER, one case had IHC-positive and polymerase chain reaction (PCR)–negative results (ER score, 4.3; H score, 128.86). For PR, six cases had negative results across all measuring systems (mean PR score, 4.06; mean H, score 20.8). Five cases were only PCR-negative (mean PR score, 4.82; mean H score, 144.95). Two cases were only H score–negative (mean PR score, 6.25; mean H score, 38.49). One case was IHC-negative and PCR-positive (PR score, 5.8; H score, 12.24). Our results were consistent with previous studies in that more IHC-positive and RT-qPCR–negative cases were observed compared to the opposite [5,7,9,10]. In concordance with earlier studies, no PR-positive, ER-negative case was found.
We reviewed the discordant cases between IHC scores and RT-qPCR scores. Regarding the IHC-positive and RT-qPCR–negative cases (one for ER and five for PR), the immunostained slides showed positivity for both the Allred score and H score (Table 5). One PR IHC-negative, RT-qPCR–positive case demonstrated a strong PR-positive ductal carcinoma in situ component within the area of hormone-negative invasive carcinoma (Fig. 5A). Under secondary review, the automatically recognized staining intensities and subsequently calculated H scores seemed accurate (Fig. 5B).
DISCUSSION
RT-qPCR methods enable quantitative and consistent measurement of clinically significant gene expression levels. In contrast, currently used manual immunohistochemical assessment systems may demonstrate a lack of reproducibility. Scoring systems that use image analyzers are expected to overcome this weakness. In this study, we found high correlations between automatically calculated immunohistochemical scores and RT-qPCR hormone expression levels.
Immunohistochemical evaluation of hormonal receptor expression status captured the tendency to oversaturation—skewed to the high expression side—, especially in the ER and a high score groups. Among all three measuring methods, the RT-qPCR score was closest to the normal distribution. Despite that the number of discordant cases for PR was higher, the correlation was higher for PR than for ER. This is because the distribution was more right-shifted in ER, resulting in a non-linear correlation with the RT-qPCR score. As the IHC score was somewhat shifted to the right, it would lose linearity as it approached a high score.
Measurements obtained with the Allred scoring system were even more right-shifted than H scores for both ER and PR [10]. More than 90% of cases had an ER Allred score of 8, and more than 50% of cases had a PR Allred score of 8. No single case had an ER Allred score less than 4. Compared to the Allred score, the H score system demonstrated a linear quantitative measurement for receptor status.
The correlation magnitude between H scores and RT-qPCR scores was not significantly different than that between Allred scores and RT-qPCR scores. ER H score showed a higher correlation coefficient compared to the Allred score in this study 0.51 vs. 0.37 (p = .121). In contrast, the PR Allred score showed a higher correlation coefficient than the H score 0.70 vs. 0.72 (p = .39). Additionally, we compared the correlation magnitude when the image analysis system calculated both the H scores and Allred scores. After excluding confounding factors, the results showed a similar tendency to the manual Allred score. Compared to the H score, the Allred score was more correlated with the RS score in both ER (0.42 vs. 0.28) (p = .14) and PR (0.50 vs. 0.43) (p = .27). When we looked into the cases of Allred score 8, computer-aided H score results showed a significant portion of moderately positive (intensity 2) nuclei as well as strong positive nuclei (intensity 3). Sometimes, moderately positive nuclei were observed more often than strong positive nuclei. The computer-recognized variation in staining intensity may have resulted in a lower correlation than the Allred score due to its complexity [19].
We reviewed the discordant cases between IHC scores and RT-qPCR scores. Regarding the IHC-positive and RT-qPCR–negative cases (one for ER and five for PR), the immunostained slides showed positivity for both the Allred score and H score. From this result, we speculated that the RT-qPCR method may have lower sensitivity compared to IHC methods in certain situations. One PR IHC-negative, RT-qPCR–positive case demonstrated a strong PR-positive ductal carcinoma in situ component within the area of hormone-negative invasive carcinoma. This intraductal component may have caused false-positive RT-qPCR results. While immunohistochemical methods detect and count only invasive tumor areas, the RT-qPCR method may incorporate intraductal components and non-tumor areas as well.
Two cases had a negative PR H score and positive Allred score and RT-qPCR score. They both had positive Allred scores under secondary review. The automatically recognized staining intensities and subsequently calculated H scores seemed accurate. These two cases had a mean H score of 38.49. We set the “score intensity” cutoff points in the image analyzing system to define nuclear staining intensities, and the cutoff points could be finely adjusted to obtain more precise results. The H score cutoff value itself (which was set at 50 in this study) can be adjusted to reduce false-negative H score results.
In our study, PR had more intermediate H score cases than ER (13/93 [13.97%] vs. 25/93 [26.88%]). As mentioned above, the intermediate group showed the lowest correlation. The intermediate group may have had more intratumoral heterogeneity and stromal influence. Intratumoral heterogeneity of PR and contaminated non-tumor areas could have caused lower RT-qPCR sensitivity compared to IHC.
Although only weak staining of more than 1% of tumor cells is a well-known cutoff value for predicting anti-hormone therapy response, the value is quite left-shifted on a percentile scale. Allred score has been assessed visually, and an inherent problem could occur because the 1% cutoff value can be arbitrary by visual assessment [20]. The therapeutic benefit of anti-hormone therapy in low ER and PR receptor groups (positivity ranges from 1% to 10%) has not yet been established. True low ER and PR groups are rare, according to some the previous reports [13,21]. More recently, some portions of this low hormonal receptor group had characteristics more like basal-like and triple-negative groups than hormone receptor-positive groups [19,22]. No low ER tumor was found in our study, while one low PR tumor case was present. The low PR (Allred score 3 [1 + 2]) case had a positive H score value (201.22), negative RT-qPCR score (4.2), and positive ER result (Allred score, 8; H score, 278.64). More careful assessment is required for these low ER and PR groups. Various methods including the RT-qPCR method and computer-aided quantification will be helpful.
In conclusion, the correlation magnitude between automated H scores and RT-qPCR scores was high and comparable to those of Allred scores. Automated H scores may become more predictive when further large-scale studies with refined methods are conducted.
The antibodies used in this study (SP1 for ER and 1E2 for PR) are well known for being more sensitive than other ER and PR antibodies, thus can reduce false-negative results [23,24]. These widely used antibodies have shown a good correlation with patient outcomes [23,24]. With the use of these antibodies, the distribution of IHC scores could be more skewed to high scores.
The Oncotype Dx test targets ER-positive, node-negative breast cancer patients only. Thus, this study was conducted only with ER-positive and high score patients, which could have led to an incomplete interpretation of the results. Further study designs that include all hormonal receptor statuses, especially low ER, PR groups (1%–10% positive cells), would be informative.
Acknowledgments
The biospecimens and data used for this study were provided by the Biobank of Korea University Guro Hospital, a member of the Korea Biobank Network. This research was supported by a grant (HI14C3396) by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare (MOHW), Republic of Korea.
Notes
Ethics Statement
The Institutional Review Board (IRB) of Korea University Guro Hospital (IRB No. 2019GR0410) approved this study. The requirement for informed consent was waived by the IRB. Investigations were conducted as per the rules of the Declaration of Helsinki of 1975, revised in 2013.
Author Contributions
Conceptualization: CK. Data curation: TJ. Formal analysis: TJ. Funding acquisition: CK, AK. Investigation: TJ. Methodology: TJ, CK. Project administration: CK. Resources: TJ, CK, AK. Supervision: CK, AK. Validation: TJ, CK. Visualization: TJ. Writing—original draft: TJ. Writing—review & editing: TJ, CK, AK. Approval of final manuscript: all authors.
Conflicts of Interest
AK, a contributing editor of the Journal of Pathology and Translational Medicine, was not involved in the editorial evaluation or decision to publish this article. All remaining authors have declared no conflicts of interest.
Funding Statement
No funding to declare.