The prognostic significance of p16 expression pattern in diffuse gliomas
Article information
Abstract
Background
CDKN2A is a tumor suppressor gene that encodes the cell cycle inhibitor protein p16. Homozygous deletion of the CDK-N2A gene has been associated with shortened survival in isocitrate dehydrogenase (IDH)–mutant gliomas. This study aimed to analyze the prognostic value of p16 and to evaluate whether p16 immunohistochemical staining could be used as a prognostic marker to replace CDKN2A genotyping in diffuse gliomas.
Methods
p16 immunohistochemistry was performed on tissue microarrays of 326 diffuse gliomas with diagnoses that reflected IDH-mutations and 1p/19q codeletion status. The results were divided into three groups (negative, focal expression, overexpression) according to the presence and degree of p16 expression. Survival analysis was performed to assess the prognostic value of p16 expression.
Results
A loss of p16 expression predicted a significantly worse outcome in all glioma patients (n = 326, p < .001), in the IDH-mutant glioma patients (n = 103, p = .010), and in the IDH-mutant astrocytoma patients (n = 73, p = .032). However, loss of p16 expression did not predict the outcome in the IDH-wildtype glioma patients (n = 223, p = .121) or in the oligodendroglial tumor patients with the IDH-mutation and 1p/19q codeletion (n = 30, p = .457). Multivariate analysis showed the association was still significant in the IDH-mutant glioma patients (p = .008; hazard ratio [HR], 2.637; 95% confidence interval [CI], 1.295 to 5.372) and in the IDH-mutant astrocytoma patients (p = .001; HR, 3.586; 95% CI, 1.649 to 7.801). Interestingly, patients who presented with tumors with p16 overexpression also had shorter survival times than did patients with tumors with p16 focal expression in the whole glioma (p < .001) and in IDH-mutant glioma groups. (p = .046).
Conclusions
This study suggests that detection of p16 expression by immunohistochemistry can be used as a useful surrogate test to predict prognosis, especially in IDH-mutant astrocytoma patients.
CDKN2A is a tumor suppressor gene located on chromosome 9p21 that encodes the cell cycle inhibitor protein p16 [1]. Genetic alterations of this gene are frequently observed in various types of human cancers [1–3]. With regard to brain tumors and prognosis, homozygous deletion of the CDKN2A gene has been reported to be associated with shortened survival in isocitrate dehydrogenase (IDH)–mutant glioma patients [4–8]. In the recent expert meeting of cIMPACT-NOW (the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy-Not Official WHO), update 5, genetic testing for the detection of CDKN2A homozygous deletion has been recommended for grade IDH-mutant astrocytic tumors. Its importance is increasingly high in adult and pediatric glioma patients [4,6,9–11].
Molecular testing to identify the CDKN2A deletion requires advanced, high-cost equipment. However, the results of molecular testing are difficult to interpret. Therefore, it is challenging to use molecular testing in routine diagnostic practice. Since the CDKN2A gene product is the p16 protein, immunohistochemical detection of p16 protein expression can be used instead of molecular testing to identify the CDKN2A gene deletion. Before 2010, in the pre-IDH-era, a few studies investigated the predictive value of p16 immunoreactivity in glioma samples [12–17]. The studies mainly targeted heterogenous groups of gliomas that did not reflect the current molecular genetics integrated classification. The sample size was small, and the results were not consistently conclusive. In addition, no large-scale comprehensive studies have been conducted on the prognostic significance of p16 immunochemistry in gliomas, particularly in this IDH-era of brain tumor diagnoses.
Meanwhile, although p16 is a tumor suppressor, aberrant over-expression of p16 protein has been observed in several tumors, including uterine cervical cancer, breast cancer, colorectal adenocarcinoma, and malignant melanoma [18–20]. In these tumors, p16 overexpression occurs by various mechanisms and has diverse prognostic implications depending on the tumor type [21]. Therefore, in addition to examining the prognostic relevance of p16 protein loss, it is also worth evaluating the prognostic implications of p16 overexpression in glioma patients.
This study aimed to determine whether p16 immunohistochemical staining can be used as a prognostic marker to replace CDKN2A genotyping in molecularly characterized diffuse gliomas. We first examined the correlation and concordance between p16 immunochemistry and CDKN2A fluorescent in situ hybridization (FISH) results. In addition, considering that p16 is a negative regulator of cell proliferation, we evaluated whether the loss of p16 expression was related to an increase in the cell proliferation marker Ki-67 labeling index. Finally, the survival analyses were performed to evaluate the prognostic implications of p16 protein expression patterns in the whole sample, and in the subgroups according to IDH-mutation and 1p/19q codeletion status.
MATERIALS AND METHODS
Subjects and data acquisition
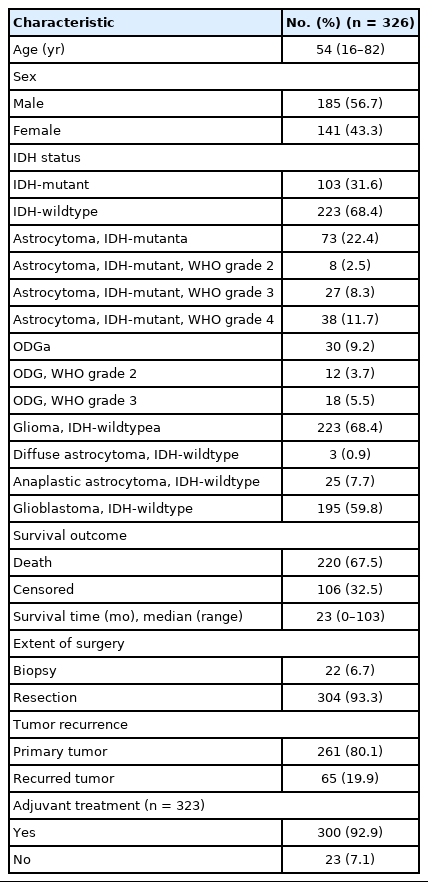
A total of 326 cases of diffuse glioma from Seoul National University Hospital (SNUH) between 2011 and 2015 with tissue microarray (TMA) blocks were included in this study. The clinical information and test results that were previously conducted to assist in diagnosis were obtained retrospectively from the electronic medical records of SNUH. The overall survival time was calculated from the date of surgery to the date of death. All diagnoses were reassessed to reflect genetic alterations, including IDH-mutation and 1p/19q codeletion status according to the cIMPACT-NOW update 5 and 6, and the 2016 World Health Organization (WHO) classification of central nervous system tumors.
Evaluation of p16 immunohistochemistry
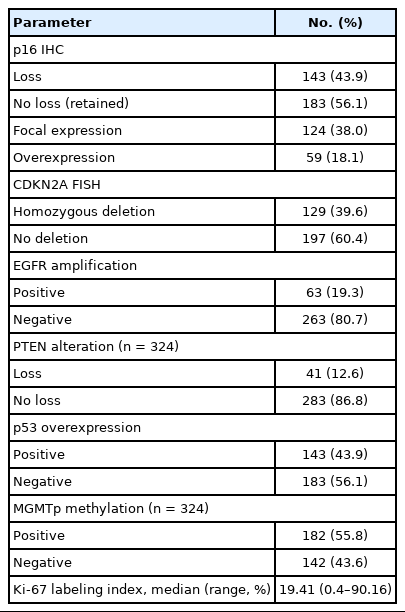
The immunohistochemical staining for p16 was conducted on the TMA sections using an antibody against p16 (mouse monoclonal, clone E6H4, Ventana, Tucson, AZ, USA) and the Ventana BenchMark XT automated immunohistochemical staining system following the manufacturer’s protocol. The results of immunohistochemical staining were first scored as the percentage of positively stained cells. Tumor cells with only nuclear or concurrent nuclear and cytoplasmic immunoreactivity were considered positive. The scoring was performed blindly without any knowledge of the diagnostic or clinical information. If there were no positive cells, or if the percentage of positive cells was < 1%, then the tumor was classified as p16 negative (loss of expression). Conversely, tumors with more than 1% immunopositivity were considered to be p16 positive (retained expression). The p16 immunopositive cases were further divided into focal expression (if the positive cells were < 50%) or diffuse overexpression (if the positive cells were > 50%) according to the degree of p16 expression. Notably, in the group with p16 overexpression, most of the tumor cells showed simultaneous intense nuclear and cytoplasmic staining (Fig. 1).
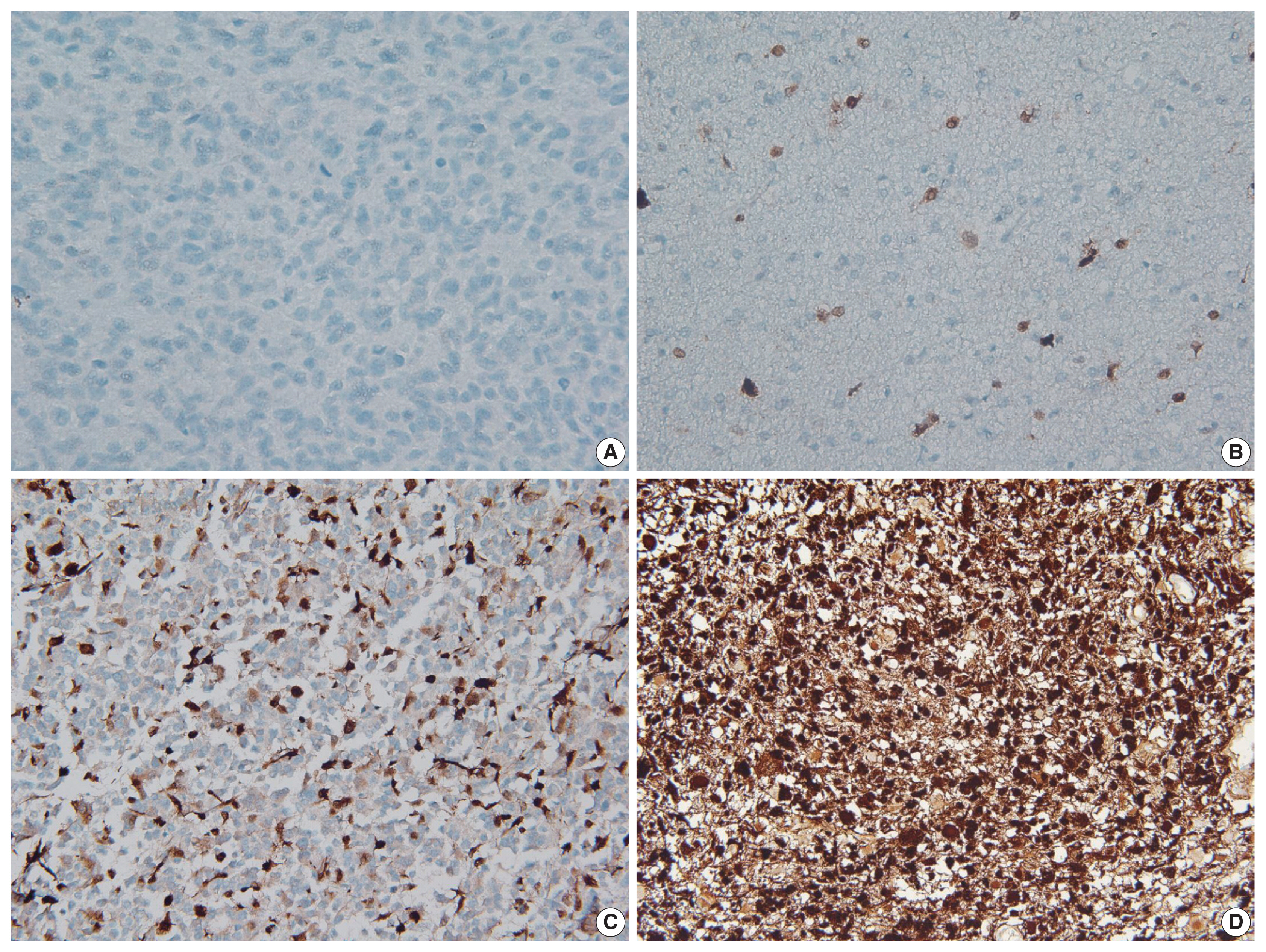
Representative images of p16 immunohistochemical staining. If immunopositive cells were absent or made up < 1% of cells, the tumor was classified as having a loss of expression (A). Conversely, tumors with > 1% of immunopositivity were considered to have retained expression, which was further subdivided into focal expression (B, C) or overexpression (D) according to the degree of p16 expression on a 50% basis. B and C show the range of focal expression.
Other immunohistochemical and molecular testing variables
The previously used antibodies included IDH1 (1:100, mouse monoclonal, clone H09, Dianova, Hamburg, Germany), p53 (1:1,000, mouse monoclonal, clone DO-7, DAKO, Glostrup, Denmark), PTEN (1:400, mouse monoclonal, clone 6H2.1, DAKO) and Ki-67 (1:100, mouse monoclonal, clone MIB-1, DAKO). Quantification of the Ki-67 labeling index was determined by the Nuclear v9 algorithm using the Aperio ImageScope software (Aperio Technologies, Vista, CA, USA). p53 was considered to be overexpressed if ≥ 30% of the tumor nuclei showed robust immunopositivity [22]. For IDH1 and PTEN, the immunohistochemical results were evaluated as positive or negative. Positive IDH1 immunostaining indicated the presence of a mutation, while negative immunostaining of PTEN meant protein loss.
Molecular tests that were previously performed for diagnosis included 1p/19q FISH to detect chromosome 1p and 19q codeletion; IDH1/IDH2 direct sequencing (performed only when the IDH1 immunohistochemistry result was negative); CDKN2A (9p21) FISH to catch gene deletion; epidermal growth factor receptor (EGFR) (7p12) FISH to detect gene amplification; PTEN (10q23) FISH to detect gene deletion; and O6-methylguanine-DNA methyltransferase (MGMT) methylation-specific polymerase chain reaction analysis. The probes used to perform FISH were as follows: Vysis LSI 1p36 SpectrumOrange/1q25 SpectrumGreen Probes and Vysis LSI 19q13 SpectrumOrange/ 19p13 SpectrumGreen Probes (Abbott Molecular, Vysis, Des Plaines, IL, USA); Vysis LSI CDKN2A SpectrumOrange/CEP 9 SpectrumGreen Probes (Abbott Molecular); Vysis LSI EGFR SpectrumOrange/CEP 7 SpectrumGreen Probes (Abbott Molecular); Vysis LSI PTEN/CEP 10 Dual Color Probe (Abbott Molecular). Each molecular test was performed and interpreted as previously described [23,24]. Homozygous deletion of the CDKN2A gene was determined when > 15% of tumor cells lost two test signals in the presence of at least one reference signal in 100 non-overlapping counted nuclei [25]. PTEN is a representative quasi-sufficient and obligate haploinsufficient tumor suppressor gene [26]. A PTEN hemizygous deletion was considered if > 50% of tumor cells showed more than one test signal loss or if > 10% of tumor cells showed two test signal losses in 100 non-overlapping nuclei with two reference signals [27]. A PTEN homozygous deletion was defined by the loss of both PTEN signals in more than 30% of tumor cells when counting more than 100 non-overlapping nuclei with at least one reference signal [27]. With regard to the PTEN evaluation, most cases were evaluated by only one of the PTEN FISH or PTEN immunohistochemistry. The final results related to PTEN loss were used in combination with the two tests.
Statistical analyses
The chi-square test was used to examine the association between categorical variables. Cohen’s kappa coefficient (κ) was calculated to determine whether there was a concordance between p16 immunochemistry and CDKN2A FISH. The Mann-Whitney U-test was used to test the relationship between p16 immunoreactivity and the Ki-67 labeling index. Kaplan-Meier survival curves were generated to estimate the overall survival distributions. The log-rank test was used for univariate survival comparisons. The Cox proportional hazards regression model was applied for the multivariate analyses using forward stepwise variable selection. Variables with prognostic significance in univariate analyses were selected as independent variables in the multivariate analysis model. Statistical significance was defined as a p-value of < .05. All statistical analyses were performed using IBM SPSS Statistics software ver. 26.0 (IBM Corp., Armonk, NY, USA).
RESULTS
Characteristics of the study population
The median age of the study population was 54 years (range, 16 to 82 years), and 185 patients (56.7%) were men. Sixty-five cases (19.9%) were of recurrent tumors. Most patients were treated with surgery, followed by adjuvant radiotherapy and/or chemotherapy. Detailed clinical information is shown in Table 1. For the survival data, the median follow-up time was 23 months (range, 0 to 103 months). During this follow-up period, 220 patients (67.5%) died, while 106 patients (32.5%) were still alive at the last contact. The diagnoses according to cIMPACT-NOW update 5 and 6 and 2016 WHO classification were as follows: astrocytoma, IDH-mutant, WHO grade 2 (n = 8, 2.5%); anaplastic astrocytoma, IDH-mutant, WHO grade 3 (n = 27, 8.3%); astrocytoma, IDH-mutant, WHO grade 4 (n = 38, 11.7%); oligodendroglioma, IDH-mutant and 1p/19q-codeleted (ODG), WHO grade 2 (n = 12, 3.7%); ODG, WHO grade 3 (n = 18, 5.5%); diffuse astrocytoma, IDH-wildtype (n = 3, 0.9%); anaplastic astrocytoma, IDH-wildtype (n = 25, 7.7%); and glioblastoma, IDH-wildtype (n = 195, 59.8%). Overall, there were 103 IDH-mutant tumors and 223 IDH-wildtype tumors. p16 loss was more prevalent in the IDH-wildtype tumors than it was in the IDH-mutant tumors (IDH-wildtype, 120 out of 223 cases [53.8%] vs. IDH-mutant, 23 out of 103 cases [22.3%]; p < .001). Among the IDH-mutant tumors, the frequency of p16 loss was similar between the IDH-mutant astrocytomas (23.3%, 17 out of 73 cases) and ODG (20%, 6 out of 30 cases) (p = .716). The age of the patients was significantly higher in the p16 loss group than it was in the p16 expression group (p16 loss group, median 56 [range, 16 to 82] vs. p16 expression group, median 50 [range, 17 to 77]; p < 0.001). There was no significant association between p16 expression status and other clinical variables such as sex, the extent of surgery (biopsy or resection), recurrence, or adjuvant treatment. The immunohistochemical and FISH results of the study population are presented in Table 2.
Comparison of p16 immunochemistry and CDKN2A FISH results
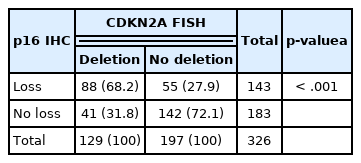
The association between the CDKN2A homozygous deletion and the loss of p16 expression was significant (p < .001) (Table 3). Sixty-eight percent (88/129) of tumors with CDKN2A homozygous deletions by FISH demonstrated a loss of p16 expression by immunohistochemistry, while 72% (142/197) of tumors without the CDKN2A homozygous deletion showed p16 immunopositivity. Therefore, the diagnostic accuracy of p16 immunochemistry to detect a CDKN2A homozygous deletion confirmed by FISH was 70.6% (250/326). There was fair agreement between the CDKN2A FISH and p16 immunohistochemistry results (Cohen’s kappa = 0.396, p < .001).
Correlation between p16 immunohistochemistry and Ki-67 labeling index
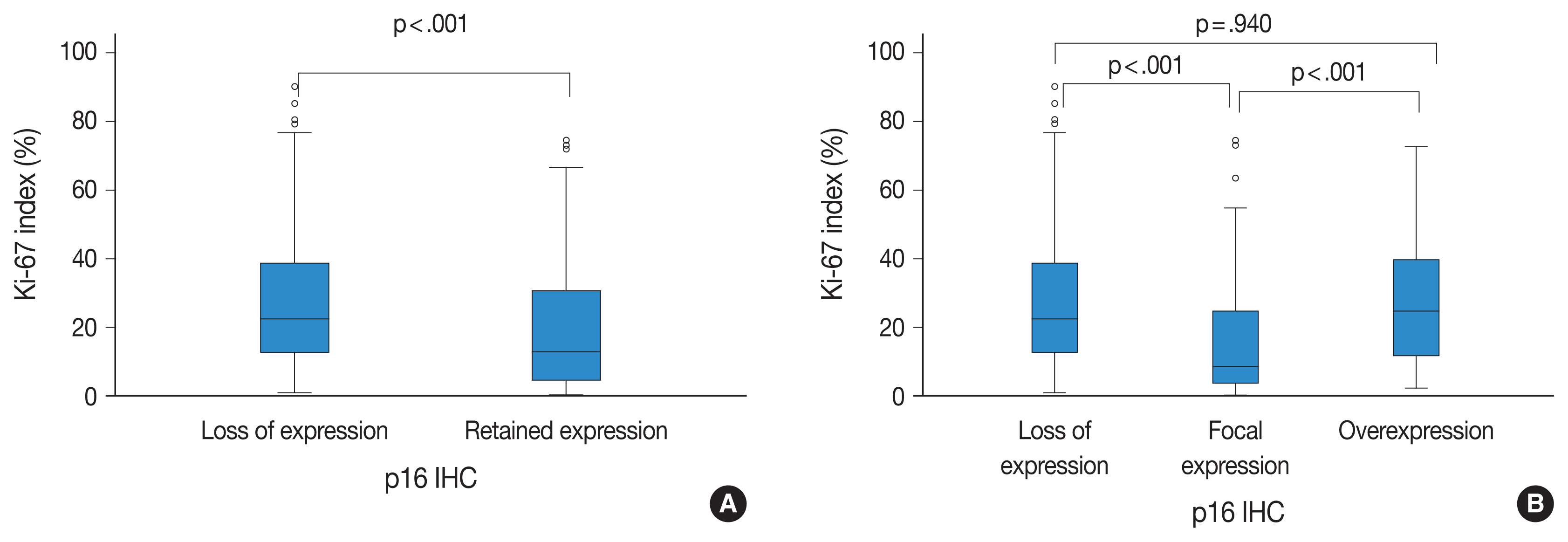
There was a significant inverse correlation between p16 expression and the Ki-67 labeling index (p < .001). That is, tumors with p16 loss had a significantly higher Ki-67 labeling index (mean, 27.60%; median, 22.50%; range, 1.11 to 90.16) than did those with p16 expression (mean, 19.98%; median, 12.90%; range, 0.40 to 74.46). In addition, when the p16 retained expression (no loss) group was divided into two additional groups of overexpression and focal expression, the p16 overexpression group (mean, 27.73%; median, 24.70%; range, 2.34 to 72.70) showed a significantly higher Ki-67 labeling index than did the p16 focal expression group (mean, 16.30%, median, 8.55%; range, 0.40 to 74.46; p < .001). There was also a significant difference in the Ki-67 labeling index between the p16 focal expression group and the p16 loss of expression group (mean, 27.60%; median, 22.50%; range, 1.11 to 90.16; p < .001). However, there was no significant difference between p16 loss of expression and the p16 overexpression groups (p = .940) (Fig. 2).
Survival analysis
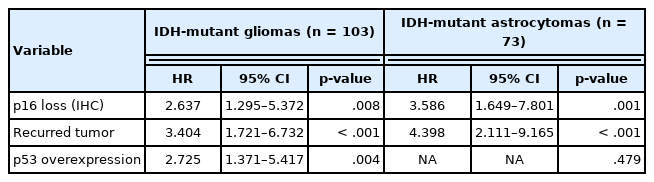
In a total of 326 whole glioma samples, a loss of p16 expression was significantly associated with short overall survival in univariate analysis using Kaplan-Meier curves and the log-rank test (p < .001) (Fig. 3A). When stratified by IDH status, tumors with p16 loss demonstrated a significantly worse outcome in IDH-mutant glioma patients (p = .010) (Fig. 3B) than did those without p16 loss. However, no such association was found in the IDH-wildtype gliomas (p = .121) (Fig. 3C). Other parameters whose prognostic significance was confirmed by the log-rank test included recurrent tumors (p < .001), EGFR amplification (p = .004) and p53 overexpression (p = .002) in IDH-mutant glioma patients. In contrast, the extent of surgery (p = .043), adjuvant treatment (p = .005) and MGMT promoter methylation (p = .024) were included in the IDH-wildtype gliomas. Sex and PTEN loss had no significant prognostic association with either group. Multivariate Cox proportional hazards regression analysis was performed on the IDH-mutant gliomas using variables selected in univariate analyses as covariates. Although CDKN2A FISH and Ki-67 were significantly prognostic for IDH-mutant gliomas in univariate analysis (p = .001, the log-rank test for CDKN2A FISH; p < .001, and univariate Cox regression analysis for Ki-67), they were not included in the final multivariate model because they were collinear with p16 immunohistochemistry. After adjusting for recurrence, EGFR amplification and p53 overexpression, p16 loss was still a significant prognostic factor for worse outcome (p = .008; hazards ratio [HR], 2.637; 95% confidence interval [CI], 1.295 to 5.372) (Table 4). Next, when the IDH-mutant group was subdivided according to 1p/19q codeletion status, loss of p16 expression was associated with significantly shorter overall survival in astrocytoma, IDH-mutant patients by the log-rank test (p = .008) (Fig. 3D), but not in ODG patients (p = .457). This association with the IDH-mutant astrocytomas was also significant in a multivariate analysis adjusted for recurrence, EGFR amplification, and p53 overexpression (p = .001; HR, 3.586; 95% CI, 1.649 to 7.801) (Table 4).
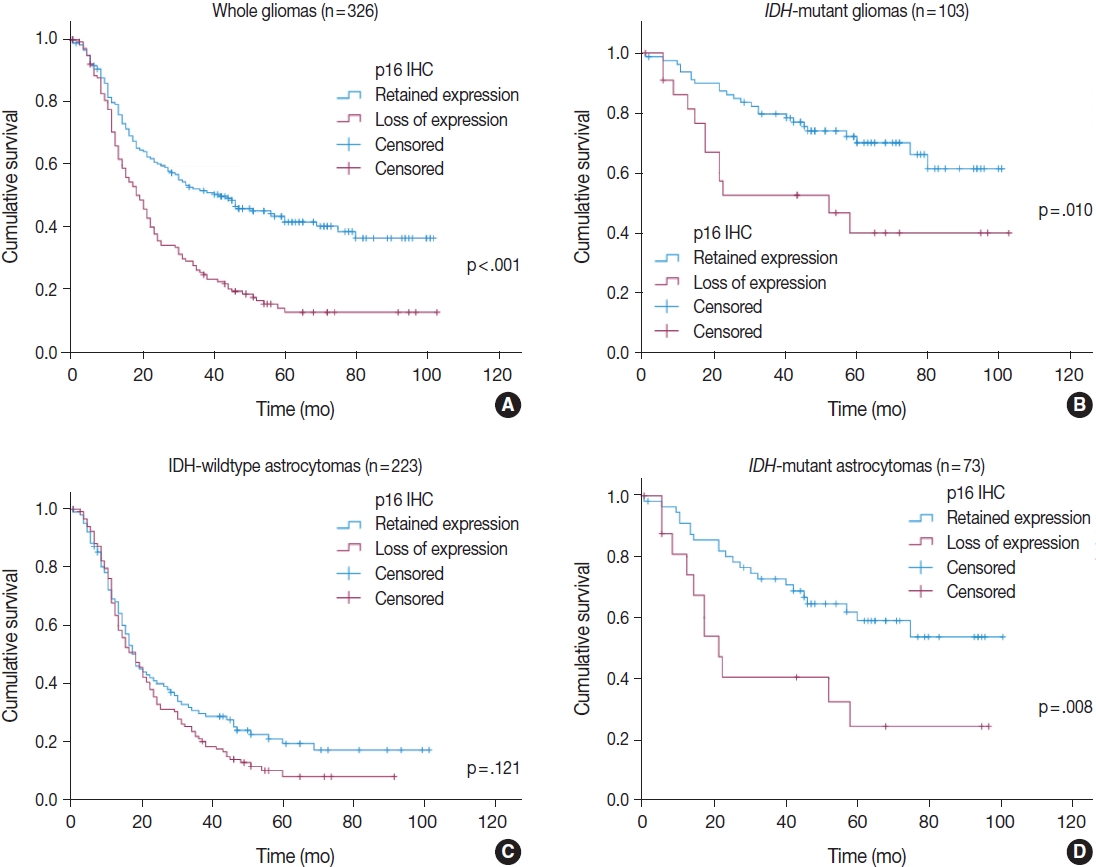
Kaplan-Meier curves for overall survival according to p16 expression status: (A) whole gliomas, (B) IDH-mutant gliomas including oligodendrogliomas, (C) IDH-wildtype astrocytomas, and (D) IDH-mutant astrocytomas. IDH, isocitrate dehydrogenase; IHC, immunohistochemistry.
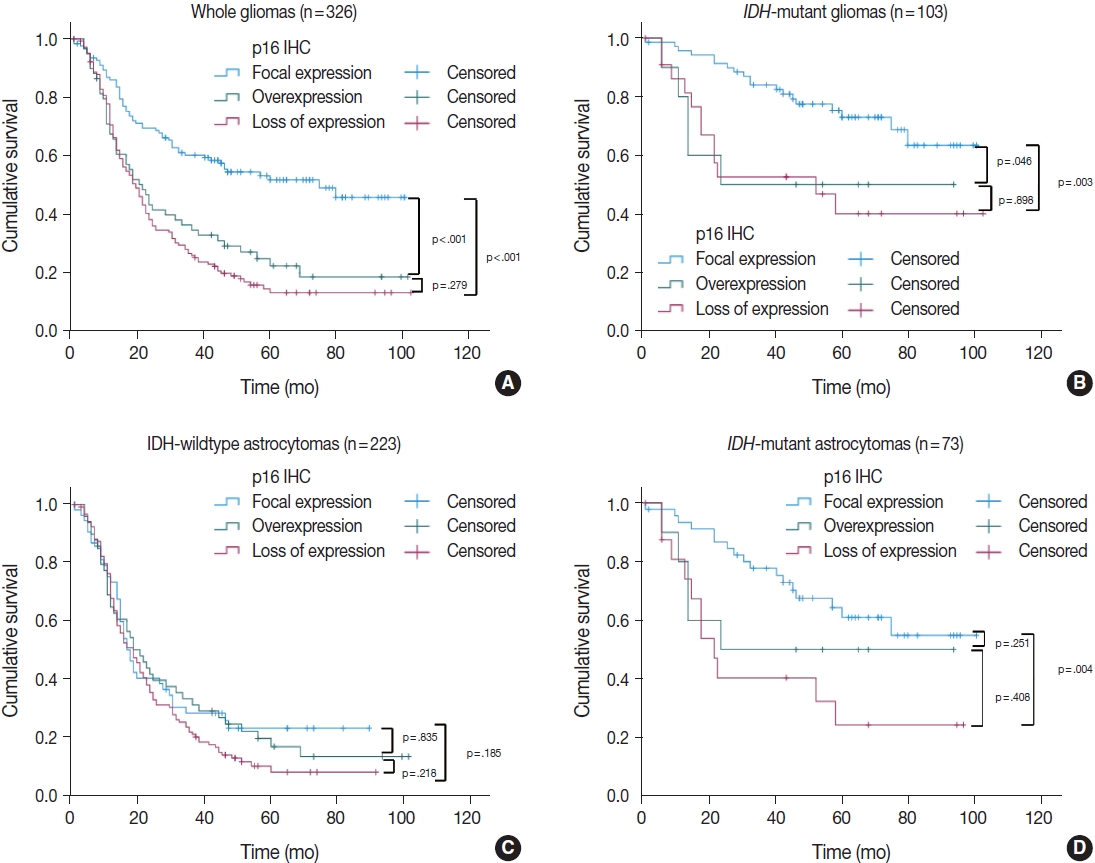
We also divided the p16 retained expression (no loss) tumors into two additional categories (focal expression vs. overexpression) according to the degree of p16 expression to check whether p16 overexpression has a prognostic meaning. Interestingly, among the 326 whole glioma cases, the p16 overexpression group showed a significantly worse overall survival curve than did the p16 focal expression group in Kaplan-Meier analysis (p < .001) (Fig. 4A). When limited to the IDH-mutant tumors (including astrocytomas and ODG), the p16 overexpression group had worse overall survival than did the p16 focal expression group (p = .046) (Fig. 4B). This association was not seen in the IDH-wildtype glioma patients (Fig. 4C). In multivariate analysis (adjusted for recurrence, p53 overexpression, and EGFR amplification), p16 overexpression was still associated with shorter overall survival than p16 focal expression, but this was not statistically significant (p = .171; HR, 2.048; 95% CI, 0.734 to 5.711). In the subgroup of IDH-mutant astrocytoma patients, p16 overexpression also appeared to be related to shorter overall survival compared to p16 focal expression; however, the difference did not reach statistical significance in Kaplan-Meier analysis (p = .251) (Fig. 4D). Meanwhile, in the ODG, the analysis was not possible because none of the 30 cases showed p16 overexpression.
DISCUSSION
This study demonstrates the prognostic value of p16 immunohistochemical staining in a large sample of molecularly characterized diffuse gliomas. Negative immunohistochemical staining of the p16 protein predicted worse overall survival in all glioma patients and in the IDH-mutant subgroup, especially in IDH-mutant astrocytomas, after adjusting for other prognostic factors such as tumor recurrence, p53 overexpression, and EGFR amplification. Our results were expected in light of existing knowledge that the p16 protein is encoded by the CDKN2A gene [1] and that CDKN2A homozygous deletion is a significant prognostic factor in IDH-mutant glioma patients [9,28]. In addition, when the p16 retained expression tumors were divided into two additional categories of overexpression and focal expression, the tumors with p16 overexpression also demonstrated worse outcomes compared to tumors with p16 focal expression in the whole gliomas and IDH-mutant gliomas including IDH-mutant astrocytomas and ODG. Considering that immunohistochemistry is a relatively simple and convenient test to be used in routine practice, assessing p16 protein expression patterns with immunohistochemical methods would be a useful way to predict glioma prognosis in the field.
Previously, several studies have examined the association between p16 immunohistochemistry and molecular tests such as PCR or FISH to detect CDKN2A deletions in glioma samples [5,13,29–31]. Some of them reported that the association was good, while others did not agree. Reis et al [5]. reported that p16 expression by immunohistochemistry correlated poorly with CDKN2A deletion by FISH, and suggested that FISH be used to evaluate CDKN2A status. This study differed from ours because the p16 immunohistochemistry data were used as continuous variables. In addition, this group counted the total number of signals regardless of whether the deletion pattern was hemizygous or homozygous when assessing the CDKN2A deletion by FISH. The problem with using the p16 expression data as a continuous variable is that it may not reflect the effect of p16 acting nonlinearly, as shown in our current study. In this study, Ki-67 was high in both the p16 loss and p16 overexpression subgroups, which was also associated with poor survival in each subgroup. The prior study appears to be the only study that has examined the relationship between p16 immunohistochemistry and survival in glioma patients molecularly diagnosed with both IDH and 1p/19q information to date. This group also found that there was a weak but significant association between p16 immunonegativity and poor overall survival in 83 astrocytoma patients.
In our study, although there was a correlation between CDKN2A FISH and p16 immunohistochemistry, the degree of agreement between the two tests was fair, or at most moderate. Therefore, our results did not show a sufficient value to suggest that substitution between the two tests is reasonable. Fifty-five out of 197 (27.9%) tumors without CDKN2A homozygous deletion (determined by FISH) demonstrated a loss of p16 expression. In contrast, 41 out of 129 (31.8%) tumors with CDKN2A homozygous deletion (by FISH) demonstrated p16 immunopositivity in the present study. Such discrepancy may come from the fact that the expression of the p16 protein is controlled not only by cytogenetic alterations, but also by other mechanisms such as point mutations or epigenetic regulations such as EZH2 mediated transcriptional repression [31]. Furthermore, immuno-positive cases with CDKN2A deletion by FISH may be due to a problem on the immunohistochemistry side, such as a hidden mixture of normal tissue. Alternatively, this result may also be due to a problem on the FISH side, such as a false-positive FISH result caused by partial hybridization failure, truncation artifacts, or a suboptimal cutoff value [32]. Similar false-positive FISH results were previously reported in another study on mesotheliomas [33], which were attributed to suboptimal hybridization of the FISH probes. False-positive FISH results may also be attributable to the heterogeneity of p16 immunostaining and CDKN2A deletions in the same tumor, as previously demonstrated in gliomas [34]. Given that FISH is relatively expensive, difficult to perform correctly, and requires a skilled technician, one must reconsider whether FISH is reliable and the gold standard of diagnosis. It is also noteworthy that p16 immunochemistry may function better as a prognostic marker than as a diagnostic one, because it reflects cases in which protein expression of the CDKN2A gene is suppressed by mechanisms other than deletion, such as epigenetic silencing or point mutations. In IDH-mutant gliomas, DNA methylation occurs frequently due to the so-called glioma CpG island methylator phenotype (G-CIMP) [35]. Detecting the absence of expressed proteins by immunohistochemistry may reflect a more ultimate situation than detecting the deletion of a gene by molecular testing.
In addition to p16 loss, we also examined whether the overexpression of p16 protein has any prognostic implications. Contrary to a cell cycle inhibitor’s original function, p16 overexpressing tumors were found to have a high proliferation index as measured by Ki-67, and to have poor prognosis, especially in IDH mutated tumors. p16 is a component of the cell cycle regulation pathway that converges into the tumor suppressor protein Rb. Disruption of Rb results in p16 overexpression in cancer tissue due to positive feedback [21]. Nakamura et al. [36] found that loss of expression of the RB1 gene was common in secondary glioblastoma. Therefore, the p16 overexpressing tumors observed in our study may be tumors that have excessively increased their p16 level to compensate for the loss of Rb. Therefore, p16 overexpression seems to be a desperate effort to stop uncontrolled proliferation due to failure of the Rb pathway. Our finding that a high Ki-67 labeling index was observed in the p16 overexpression group is considered to be in good agreement with this situation. Therefore, for p16 overexpressing tumors, we must determine whether there is another genetic abnormality in the Rb pathway components that include RB1 deletion or CDK4/6 amplification, and whether this is associated with prognosis.
In the previous literature, there were inconsistent findings on whether a CDKN2A homozygous deletion was associated with poor survival in IDH-mutant and 1p/19q-codeleted ODGs [4,8, 28]. Similarly, our study did not show any prognostic significance of p16 protein loss in ODG. However, our results must be interpreted with caution given the small sample of ODG cases and a relatively short follow-up period for lower-grade gliomas. Therefore, further research is likely needed. It was also significant that p16 overexpression was not observed in all of our study’s 30 ODGs. More samples are needed to confirm that the ODGs do not have p16 overexpression.
A limitation of this study is that it was retrospective in nature. Therefore, the data collection was inevitably limited, and other factors that may be related to prognosis were not all included or excluded. Most importantly, the FISH data should be reevaluated. We were not able to review these data because the preserved FISH pictures were limited. In addition, we cannot rule out the possibility that the immunohistochemical readings were overestimated or underestimated if the tumor had heterogeneous p16 expression, because the analysis was performed using TMA slides. Meanwhile, p16 overexpression was a statistically significant prognostic factor, which was confirmed in univariate analysis of IDH-mutant gliomas including astrocytomas and ODG. This significance was concealed after multivariate analysis or after being divided into subgroups, which may be due to insufficient sample sizes and short follow-up. Therefore, larger studies are necessary.
In summary, this study demonstrated that the pattern of p16 expression was significantly correlated with the prognosis in IDH-mutant glioma patients. p16 immunohistochemistry was correlated with CDKN2A FISH. The loss of p16 expression was strongly associated with shortened overall survival. In addition, the overexpression of p16 was also related to a worse outcome. We suggest that detecting p16 protein expression by immunohistochemistry could be used as a useful surrogate test or an initial screening assay to predict patient prognosis while replacing CDKN2A genetic testing. Nevertheless, further studies in other cohorts may be required to confirm these results.
Notes
Ethics Statement
All procedures performed in the current study were approved by the Seoul National University Hospital Institutional Review Board (IRB No. H-2008-111-1150) in accordance with the 1964 Helsinki declaration and its amendments. Formal written informed was waived for cases collected before February 2013. Otherwise, informed consent was obtained from each patient.
Author Contributions
Conceptualization: SHP, JWP. Data curation: JWP, SHP. Formal analysis: JWP. Funding acquisition: SHP. Investigation: JWP, JK, KYL, HK, SIK, JKW, SHP. Methodology: JWP, SHP. Resources: CKP. Supervision: SHP. Writing—original draft: JWP. Writing—review & editing: JWP, SHP, JKW. Approval of final manuscript: all authors.
Conflicts of Interest
The authors declare that they have no potential conflicts of interest.
Funding Statement
This study was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHI-DI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C1277).