Fusobacterium nucleatum: caution with interpreting historical patient sample cohort
Article information
Fusobacterium nucleatum (Fn) was first noted to be associated with colorectal cancer (CRC) in 2012 [1]. Since then there have been several publications in retrospective cohorts analyzing the relationship between colorectal tumor fusobacterial abundance, clinical and molecular characteristics and clinical outcomes [1]. The majority of studies, including ‘Prognostic impact of Fusobacterium nucleatum depends on combined tumor location and microsatellite instability status in stage II/III colorectal cancers treated with adjuvant chemotherapy’ by Oh et al. [2] published in Journal of Pathology amd Translational Medicine, utilize quantative polymerase chain reaction for the NusG gene in Fn relative to a control prostaglandin transporter gene (SLCO2A1). Many of these studies utilize formalin-fixed paraffin-embedded (FFPE) tissue that has been stored for up to two decades. Although this enables a long follow-up period for clinical outcomes, recent data from our lab shows that advanced age of sample significantly impairs the ability to detect Fn.
Oh et al. [2] analyzed 747 surgical samples of stage III and high-risk stage II CRCs that had been preserved in FFPE format. These participants had surgery between 2005 and 2012—making their specimens 6–13 years old at time of analysis. Five hundred and ninety-three samples were of quality to be included in the analysis. Oh et al. [2] noted a detection rate for Fn of 68.8%. They used a median cutoff to divide Fn-positive results into Fn-high and -low then grouped Fn-low with Fn-negative for analysis. Oh et al. [2] found participants with Fn-high CRCs to have significantly higher T score and were more likely to be proximal cancers. Fn-high non-sigmoid colon cancers that subsequently received adjuvant chemotherapy had significantly improved survival compared to Fn-low/negative. This association was intensified among the MSI-low sub-group.
In their discussion, Oh et al. [2] make note of their unpublished data showing more recently collected and embedded specimens had higher rates of Fn positivity. They infer Fn detection rate could be affected by storage time.
Based on our data, we would like to support this unpublished finding of Oh et al. [2] and demonstrate a significant negative relationship between storage time of FFPE tissue and detection of Fn.
We analyzed fusobacterial abundance in diagnostic biopsies from 202 locally advanced rectal cancers (LARCs) from patients diagnosed and treated in Irish hospitals between 2000 and 2020, using the same qPCR method as Oh et al. [2].
Ten point four percent of LARC diagnostic biopsies were positive for Fn. The age of sample at time of analysis was measured in months between diagnostic biopsy and analysis by qPCR. Positive samples were more likely to be younger. The median sample age among Fn-positive samples was 57 months, compared to 134 months for Fn-negative samples. The age of samples was significantly different between negative and positive groups on Wilcoxon Rank-sum (z = 3.568, p < .001) (Fig. 1).
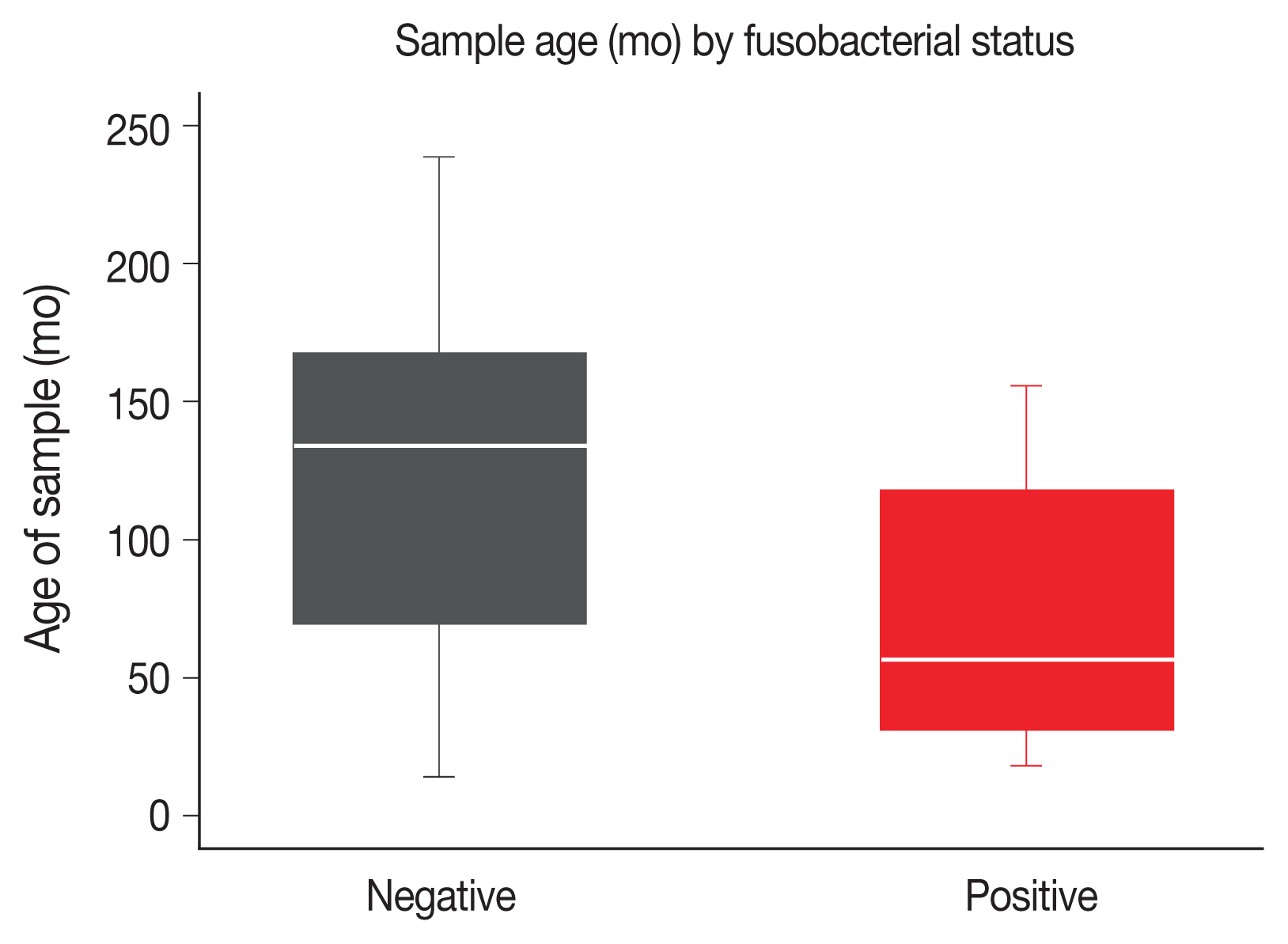
Age of sample at time of analysis in months by fusobacterial status (negative for Fn vs. positive for Fn). The negative samples are significantly older than the positive samples (median age, 134 months vs. 57 months; p < .001 Wilcoxon rank-sum).
The samples in our study, and that by Oh et al. [2], were preserved in FFPE format. FFPE processing can induce DNA damage itself and over time DNA slowly degrades in FFPE format and length of amplifiable DNA fragments become shorter [3,4]. Most research on DNA degradation focuses on mammalian, eukaryotic DNA but there is emerging evidence that FFPE preservation may disproportionately affect bacterial DNA [4,5].
A review of published studies detecting Fn with the same qPCR method with change in cycle thresholds (qPCR ΔCT) shows some tendency for older sample cohorts to have lower rates of Fn detection. Table 1 compares rates of Fn detection from lowest to highest between publications that used similar methods to evaluate Fn in CRC tumor samples, format samples were stored in and the age of the samples at time of analysis. Sample age was calculated from the difference between years of collection and date of publication, but it is likely DNA may have been extracted years before analysis and that analysis may have occurred sometime before publication in some cases [6–27]. Only Oh et al. [2] and Mima et al. [8–10] commented that year of diagnosis (and therefore age of sample) was associated with Fn detection rates. There appears to be a pattern between age of cohorts and rates of detection of Fn, but frozen samples and fluorescent in situ hybridization or ddPCR method appear to increase sensitivity too. Unfortunately, as studies did not include supplementary data or reports of age of samples meta-analysis was not possible, but we acknowledge this would be ideal.
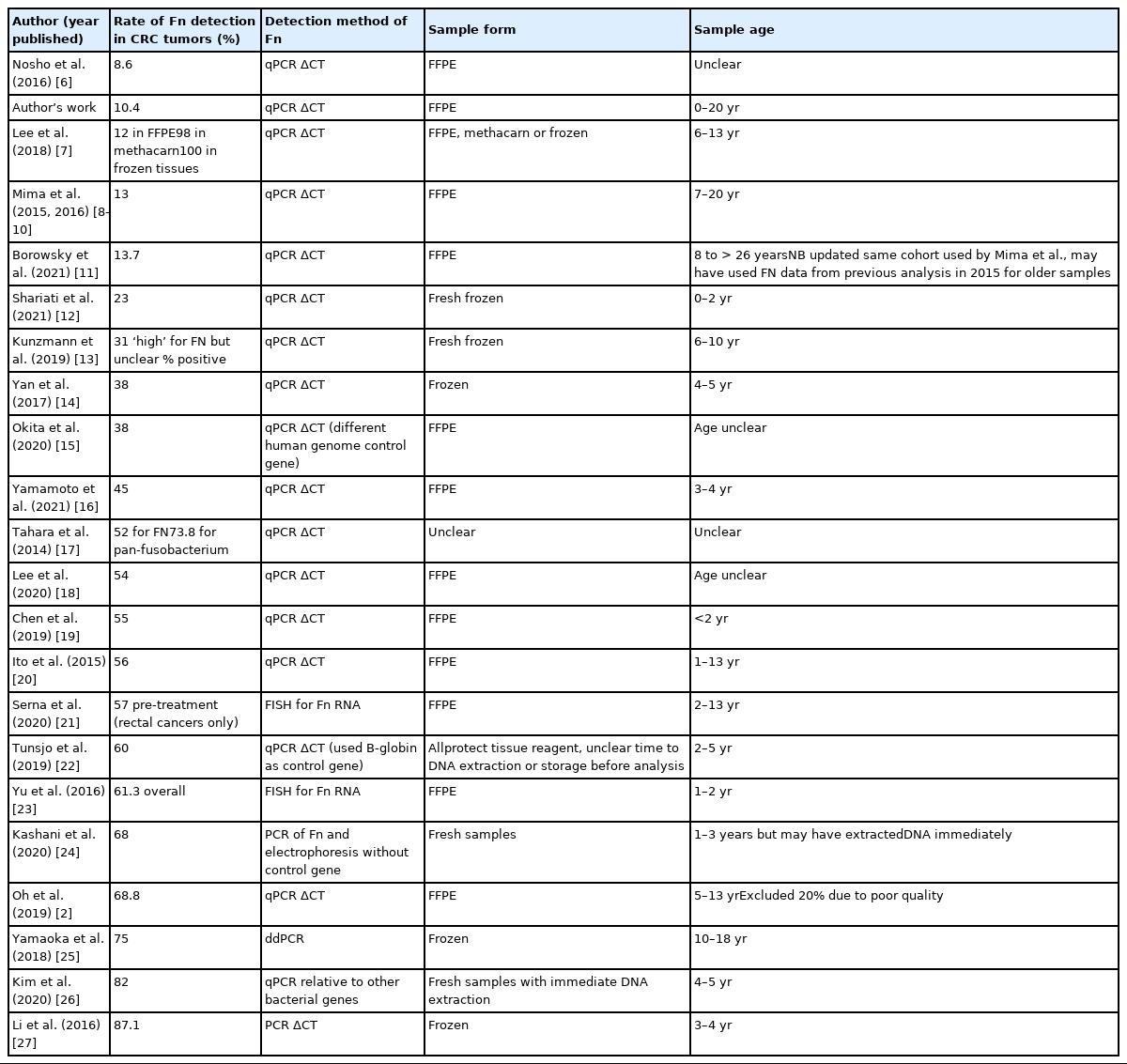
Comparison of Fn detection rates in publications analyzing CRC tumors by detection method, sample storage format, and sample age
As shown by us and Oh et al. [2], the ability to detect Fn from FFPE CRC tissue using qPCR declines significantly with time. Even with adjustment for age of sample, the association between Fn-status and prognosis (or other clinical outcomes) may be underestimated by using older cohorts with FFPE specimens. This is important to bear in mind for interpretation of previously published retrospective cohorts and suggests that more recently diagnosed cohorts may be more appropriate to use in the future.
Notes
Ethics Statement
Tumor samples were obtained from patients enrolled on the TRI-LARC clinical trial (NCT02151019). Historical tumor samples were obtained from tumor banks under the auspices of Institutional Review Board-approved protocols. Informed consent for tumor biobanking and research studies was obtained from all patients.
Availability of Data and Material
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
Author Contributions
Conceptualization: KLFJ, ST, BTH. Data curation: KLFJ, ST. Formal analysis: KLFJ, SM. Funding acquisition: ST, BTH, BDPO. Investigation: KLFJ, ST. Methodology: KLFJ, ST. Supervision: ST, BTH. Validation: KLFJ, ST. Writing—original draft: KLFJ. Writing—review & editing: KLFJ, ST, SM, BTH. Approval of final manuscript: all authors.
Conflicts of Interest
The authors declare that they have no potential conflicts of interest.
Funding Statement
No funding to declare.
