Evaluation of Protein Expression in Housekeeping Genes across Multiple Tissues in Rats
Article information
Abstract
Background
Housekeeping genes, which show constant protein expression patterns between different tissue types, are very important in molecular biological studies as an internal control for protein research.
Methods
The protein expression profiles of seven housekeeping genes (HPRT1, PPIA, GYS1, TBP, YWHAZ, GAPDH and ACTB) in various rat tissues (cerebrum, cerebellum, cardiac ventricle and atrium, psoas muscle, femoral muscle, liver, spleen, kidney, and aorta) were analyzed by Western blot and compared by coefficient of variation (CV).
Results
HPRT1 was stably expressed (CV≤10%) in six tissues (cerebrum, cerebellum, ventricle, femoral muscle, spleen, and kidney), PPIA was stably expressed in five tissues (cerebrum, cerebellum, ventricle, spleen and kidney), YWHAZ was stably expressed in three tissues (cerebrum, cerebellum, and kidney), and GAPDH was stably expressed in four tissues (cerebrum, ventricle, psoas muscle, and kidney). In comparison, GYS1, TBP, and ACTB were found to have CV values over 10% in all tissues. Of the seven genes examined, four (HPRT1, PPIA, YWHAZ, and GAPDH) were found to be stably expressed across multiple organs, with low CV values (≤10%).
Conclusions
These results will provide fundamental information regarding internal controls for protein expression studies and can be used for analysis of postmortem protein degradation patterns in forensic medicine.
In molecular biological studies on changes in mRNA and protein expression, it is necessary to establish an internal standard set of genes that do not significantly change with different conditions or study objects. Housekeeping genes are essential genes that maintain basic cellular functions. They are expected to be consistently expressed in all cells, not only under a normal state, but also under diseased conditions. Nevertheless, some housekeeping genes do change their expression with diverse conditions, including cultured cell density or injurious conditions.1,2 Different sets of housekeeping genes are used for various purposes in molecular biology, and many reference genes have been considered as a standard in gene expression studies. Most previous studies related endogenous control genes to mRNA expression using quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR). Through a review of 370 published papers (NCBI-Pubmed, as of January 2010) analyzing swine mRNA expression, Gu et al.3 showed that the most widely used endogenous housekeeping genes were ACTB (30.62%), GAPDH (27.24%), and 18S (12.52%). The researchers recommended the three most stable housekeeping genes in all analyzed samples (TOP2B, HSPCB, and YWHAZ) for accurate normalization of qRT-PCR data.3 They also suggested that different sets of housekeeping genes were appropriate for adipose tissue (HSPCB, ALDOA, and GAPDH genes) and for muscle tissue (TOP2B, HSPCB, and YWHAZ genes).3 According to the study by Zuo et al.,4 RT-PCR results revealed that the porcine GYS1 gene was expressed in the spleen, lung, liver, kidney, small intestine, skeletal muscle, heart, and stomach, with the highest expression level in skeletal muscle.
In order to define housekeeping genes based on the aforementioned facts, the expression stabilities of seven housekeeping gene-coded proteins-hypoxanthine phosphoribosyl-transferase I (HPRT1), peptidylprolyl-isomerase A (PPIA), glycogen synthase 1 (GYS1), TATA box binding protein (TBP), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta polypeptide (YWHAZ), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and beta-actin (ACTB)-were evaluated using various rat tissues including cerebrum, cerebellum, cardiac ventricle and atrium, psoas muscle, femoral muscle, liver, spleen, kidney, and aorta. By examining these seven housekeeping genes, this study aimed to identify the genes that show consistently stable expression across multiple tissues, and to provide baseline data on protein degradation patterns of internal control genes for further study in the estimation of postmortem intervals.
MATERIALS AND METHODS
Animals
A total of eight, eight-week-old male Sprague-Dawley rats were used in this study. The rats were sacrificed by decapitation under anesthesia. Then the tissues from the cerebrum, cerebellum, left ventricular free wall, whole atrial tissue, psoas muscle, femoral muscle, liver, spleen, kidney, and aorta were immediately collected and divided into several small pieces. Tissue samples were put into 1.5 mL tubes and stored at -80℃. This study was approved by the institutional Animal Ethics Committee of Chonnam National University Medical School.
Protein extraction and Western blot analysis
PRO-PREP Protein Extraction Solution (iNtRON Biotechnology, Seongnam, Korea) was added to the 1.5 mL tubes containing frozen samples according to the manufacturer's protocol. The tissues were chopped broken into small pieces using microscissors on ice for 30 minutes. The samples were centrifuged at 14,000×g and protein-containing supernatant was moved into a new tube. The protein concentration was measured using a bicinchoninic acid protein assay (Thermo Scientific, Rockford, IL, USA). Twenty microliters samples containing 10 µg total protein were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and subsequently transferred to a nitrocellulose membrane (PALL, Port Washington, NY, USA) by electrophoresis.
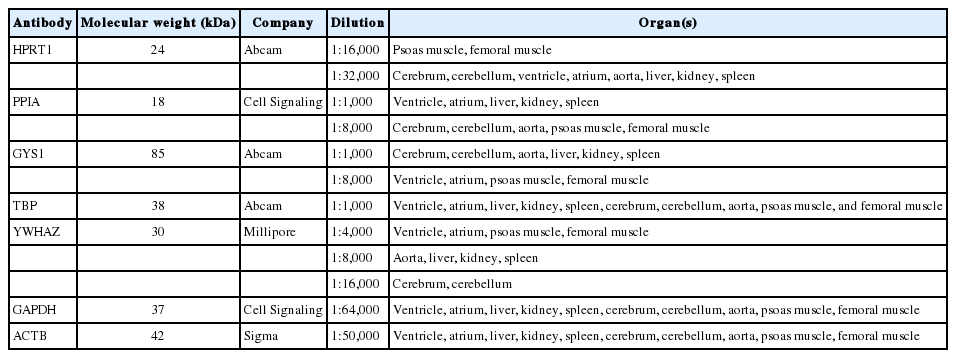
After treatment with 5% nonfat milk in Tris-buffered saline and Tween 20 (TBST) for one hour at room temperature, the membranes were incubated with primary antibodies (Table 1) overnight at 4℃. The membranes were then washed three times in TBST and incubated with horseradish peroxidase-conjugated secondary antibodies (1:10,000) for one hour. Finally, the signals were developed using an enhanced chemiluminescence kit (Millipore, Billerica, MA, USA) and analyzed using a LAS3000 (Fuji Film, Tokyo, Japan).
Quantitation and comparison of signal intensity
Points of interest (POIs) from Western blot bands were marked and quantitated using FUJI FILM MultiGauge V3.0 software (Fjji Film), and the background level was subtracted from the POI level. The data were presented as mean±standard deviation (SD) values. In addition, coefficient of variation (CV) was used for comparing the expression stability of genes in various organs. CV is defined as the SD divided by the mean, multiplied by 100%.2,5 The expression stability was evaluated based on the assumption that a lower CV value means lower variations of POI expression levels within each group. CV values equal to or less than 10% were used for selecting stable genes in a previous study,6 the same criterion was adopted in this study.
RESULTS
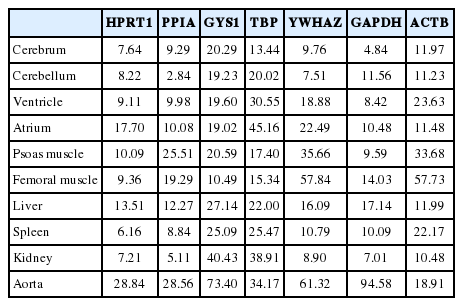
All seven housekeeping genes used in this study were expressed in all ten different tissues (Fig. 1). The signal intensities of POI measured by MultiGauge V3.0 are shown in Table 2. The CV value was calculated using the mean and SD, and the result was rounded off to two decimal places (Table 3). The expression stability of each housekeeping gene was represented using CV values across tissue types (Fig. 2). The unit of the vertical axis for CV values was expressed on a logarithmic scale to represent 10% or lower differences in each gene effectively. Among the ten organs, the cerebrum, cerebellum, ventricle, femoral muscle spleen, and kidney were shown to have stable HPRT1 expression by low CV values (≤10%); the cerebrum, cerebellum, ventricle, kidney, and spleen showed stable PPIA expression; the cerebrum, cerebellum, and kidney showed stable YWHAZ expression; and the cerebrum, ventricle, psoas muscle, and kidney, showed stable GAPDH expression. All organ tissues were found to have CV values over 10% for GYS1, TBP, and ACTB. Of the various organs examined, the femoral muscle showed the lowest CV value with GYS1. Similarly, the cerebrum showed the lowest CV value with TBP, while the kidney showed the lowest variation with ACTB.
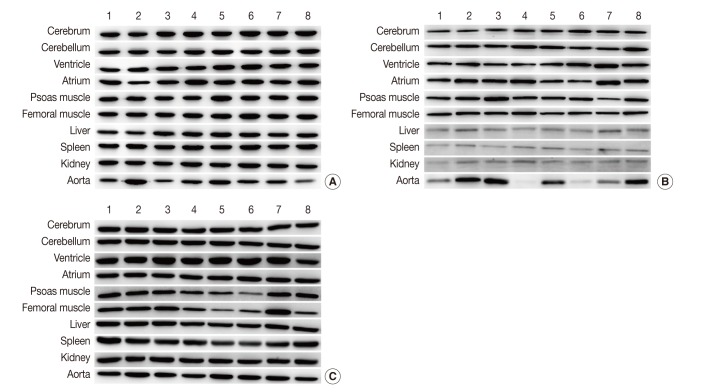
Representative photos of Western blot analysis. (A) HPRT1 exhibits reproducibility in the cerebrum, cerebellum, spleen, but shows notable variation in the aorta. (B) GYS1 exhibits reproducibility in the cerebrum, cerebellum and femoral muscle, but it shows only faint bands in the liver, spleen, and kidney. (C) ACTB exhibits relatively reproducible expression level in the cerebrum, cerebellum, atrium, liver and kidney. Unlike other housekeeping genes, ACTB exhibits reproducibility in the aorta (individual rat numbers are shown as case no. 1 through no. 8).

Average signal intensities in eight rats for comparison of protein expression in each gene across tissue types
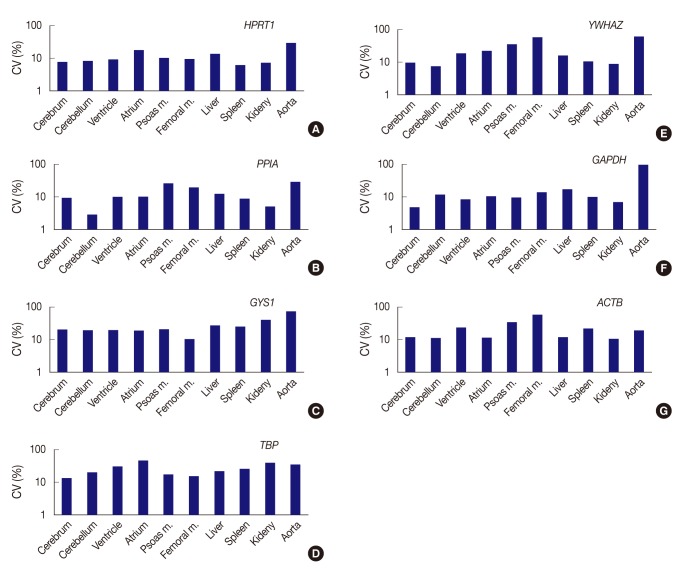
Expression stability of each gene represents as coefficient of variation (CV) value across tissue types. (A) HPRT1 shows stability in order of the spleen, kidney, cerebrum, cerebellum, ventricle, and femoral muscle, but a remarkably high CV value in the aorta. (B) PPIA shows the highest stability in the cerebellum. (C) With GYS1, all organ tissues are found to have CV values over 10%. The femoral muscle is relatively stable compared to other genes. (D) With TBP, all organ tissues are found to have CV values over 10%. (E) YWHAZ exhibits the highest stability in cerebellum, whereas it shows prominently high CV values in the femoral muscle and aorta. (F) GAPDH demonstrates the highest stability in the cerebrum, but significant variance in the aorta. (G) With ACTB, all organ tissues are found to have CV values over 10%. However, the kidney is relatively stable compared to other genes. Femoral m., femoral muscle; psoas m., Psoas muscle.
The expression stability of POI in each tissue was also evaluated using CV values, with the horizontal axis representing each gene and the vertical axis for the CV value on a logarithmic scale (Fig. 3). Among the seven genes, HPRT1, PPIA, YWHAZ, and GAPDH were shown to be stably expressed in the cerebrum with low CV values (≤10%); HPRT1, PPIA, and YWHAZ were stably expressed in the cerebellum; HPRT1, PPIA, and GAPDH, were stably expressed in the ventricle; GAPDH, was stably expressed in psoas muscle; HPRT1, was stably expressed in femoral muscle; HPRT1 and PPIA were stably expressed in the spleen; and HPRT1, PPIA, YWHAZ, and GAPDH, were stably expressed in the kidney. In the atrium, liver, and aorta, all genes were found to have CV values over 10%. Of the seven genes, PPIA showed the lowest CV value in the atrium while ACTB had the lowest CV values in the liver and aorta.
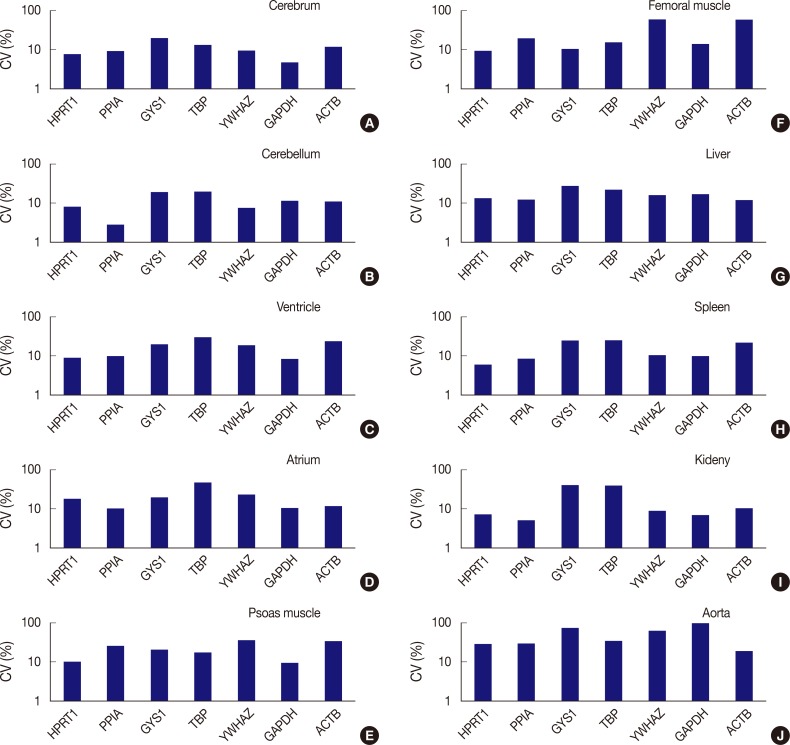
Comparison of expression stability between genes in each tissue type. (A) In the cerebrum, GAPDH demonstrates the highest stability, while GYS1 shows the least stable expression. (B) In the cerebellum, PPIA shows the highest stability, while TBP was the least stable. (C) In the ventricle, GAPDH exhibits the highest stability, while TBP was the least stable. (D) In the atrium, all seven genes are found to have coefficient of variation (CV) values over 10%. However, PPIA is relatively stable compared to other genes. (E) In psoas muscle, GAPDH shows the highest stability. (F) In femoral muscle, HPRT1 exhibits the highest stability. YWHAZ and ACTB demonstrate the highest variability in the femoral muscle. (G) In the liver, all genes are found to have CV values over 10%. However, ACTB is relatively stable compared to other genes. (H) In the spleen, HPRT1 demonstrates the highest stability. (I) In the kidney, PPIA shows the highest stability. (J) In aorta, all seven genes demonstrate high CV values over 10%.
In summary, HPRT1 in the cerebrum, cerebellum, ventricle, femoral muscle, spleen, and kidney, PPIA in the cerebrum, cerebellum, ventricle, spleen, and kidney, YWHAZ in the cerebrum, cerebellum, and kidney, and GAPDH in the cerebrum, ventricle, psoas muscle, and kidney showed 10% or lower CV values. Of the ten organs used in the study, the atrium, liver, and aorta showed high CV values greater than 10% with all housekeeping genes. Therefore, these three organs appear to be less useful tissues for studies based on constant gene expression.
DISCUSSION
In protein expression studies, it is important to identify housekeeping genes that show comparatively stable expression patterns across body tissues. Expression of certain housekeeping genes can vary widely depending on different tissue types and disease processes involved. Suitable housekeeping genes as an internal control under certain defined conditions have been reported in previous studies. Rho et al.7 have reported that RPL29 only or the combination of RPL29 and B2M was most appropriate for stomach tissues; B2M only or the combination of B2M and GAPDH for stomach cancer cell lines;7 ACTB and SDHA, for the gene expression analysis of breast cancer among typical reference genes;8 and GAPDH, RPS17, RPL30, RPLP0, and TFRC, in studies on post-mortem brain samples from patients with major psychiatric disorders.9 According to gene expression studies of mice myocardial infarction models, HPRT, RPL13A, and TPT1 are the appropriate reference genes;10 GUSB, PPIA, and TBP, are used for serous ovarian cancer;11 and EEF1A1 only or the combination of EEFA1 and GAPDH, are commonly used for cervical cancer.12 In swine skeletal muscles, PPIA, HPRT, and eEF-1γ are the genes that showed the most stable expression.13 In the rat hippocampus after resuscitation from a cardiac arrest, a combination of CypA and Pgk1 should be considered at four and 21 days after injury, whereas CypA and Gapdh is the best combination at seven days.14 CrypA is most favorable if restriction to a single reference gene is required for all time points.14 These stable housekeeping genes were identified based on mRNA expression analyses. However, in the present study, we identified the stable housekeeping genes based on protein expression.
The expression stabilities of seven housekeeping genes (HPRT1, PPIA, GYS1, TBP, YWHAZ, GAPDH, and ACTB) in the rat cerebrum, cerebellum, cardiac ventricle and atrium, psoas major muscle, femoral muscle, liver, spleen, kidney, and aorta tissues were evaluated through protein quantitation, Western blotting, and Multi-gauge software (Fuji Film). The results revealed that four genes-HPRT1, PPIA, YWHAZ, and GAPDH-showed 10% or lower CV values, which implied that they are more stably expressed across tissues compared to other housekeeping genes. The CV is one of the most used statistical measures by researchers in the evaluation of experimental precision. In field experiments, if the CV is below 10%, it is considered low. CV values from 10% to 20% are considered moderate, implying good precision, while those from 20% to 30% are considered high, indicating low precision. Finally, CV values above 30% are considered very high, indicating very low precision.15,16 Listed in order of stability, HPRT1 is a stably expressed gene in the spleen, kidney, cerebrum, cerebellum, ventricle, and femoral muscle; PPIA is stably expressed in the cerebellum, kidney, spleen, cerebrum, and ventricle; YWHAZ, is stably expressed in the cerebellum, kidney, and cerebrum; and GAPDH is stably expressed in the cerebrum, kidney, ventricle, and psoas muscle. In particular, the gene that showed the most stable expression was PPIA in the cerebellum. For GYS1, TBP, and ACTB, all organ tissues were found to have CV values over 10%. Furthermore, among the ten selected organs, the atrium, liver, and aorta were considered as less desirable tissues for housekeeping gene expression studies because of unstable expression with CV values over 10%. In studies on muscle samples, different results have been reported. Gu et al.3 reported TOP2B, HSPCB, and YWHAZ genes were appropriate for five types of muscle tissue (diaphragm muscle, heart, longissimus dorsi muscle, psoas major muscle, and tongue). In swine skeletal muscles, PPIA, HPRT, and eEF-1γ were considered to be the most stable genes.13 However, in this study, YWHAZ was not a suitable gene in the cardiac ventricle, atrium, psoas muscle, and femoral muscle, and PPIA was not stable in femoral muscle. These differences are thought to stem from differences in the protocols used.
The internal organs are protected by the skin and soft tissues from the environment, so they are less affected by external factors. However, the degradation of body organ proteins is also influenced by temperature or pH. In skeletal muscle, some proteins showed a temperature-dependent cleavage over the first 48 hours postmortem. For example, the transformation of calcineurin A was completed within 24 hours. In contrast, protein phosphatase 2A increased within the first 24 hours after which it degraded at 21℃, but remained stable for up to 96 hours at 5℃ and 10℃.17 In forensic medicine, further studies could define appropriate correction factors that would be applied to compensate for the effect of temperature changes due to environmental conditions such as clothing, water suspension, leaf and other coverings, air movement, and surface properties.18 Melody et al.19 suggested that there might be an important linkage between the intercellular environment in the postmortem skeletal muscle and the activation of µ-calpain. Moreover, the rate of postmortem pH decline within the first six hours after death was shown to influence the rate of µ-calpain activity and autolysis that plays a pivotal role in regulating early postmortem proteolysis.19 Tissue accessibility, postmortem stability, and asepsis are all important attributes for postmortem interval changes, which are afforded by some tissues (e.g., muscle), but not by others (e.g., lung).17
Based on previous studies, we searched more reference studies on postmortem proteolysis associated with postmortem intervals, but even data of protein expression on housekeeping genes were very rare. Once the pattern of postmortem protein degradation in the body is established, it can be used to estimate postmortem intervals, because some housekeeping genes analyzed in this study were stable in certain tissues. Since some animal and human proteins share high sequence homology, it is likely that the results from animal experiments will reflect human postmortem changes.17 In order to compare protein degradation during the postmortem interval, housekeeping genes that do not show differences in expression among individuals before death are needed as references, regardless of the damage or disease that caused the death. The current study is believed to offer a basic foundation for identification of proper housekeeping genes that would be selected for internal controls across various tissue types during postmortem intervals.
As mentioned earlier, almost all other studies have been performed under diseased states. In contrast, the present study may help select stable housekeeping genes from normal body tissues other than the tissues under an altered condition or disease. Baseline data for further study on protein degradation patterns of internal control genes were presented in the current study. Accordingly, the results of this study may be used for determination of the elapsed time after death. However, in order to quantitate the patterns of postmortem protein degradation, other factors including temperature or pH should be considered in future studies.
Acknowledgments
This study was supported by a Korea Basic Science Institute NAP grant (T33780).
Notes
No potential conflict of interest relevant to this article was reported.