A Multi-institutional Study of Prevalence and Clinicopathologic Features of Non-invasive Follicular Thyroid Neoplasm with Papillary-like Nuclear Features (NIFTP) in Korea
Article information
Abstract
Background
In the present multi-institutional study, the prevalence and clinicopathologic characteristics of non-invasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP) were evaluated among Korean patients who underwent thyroidectomy for papillary thyroid carcinoma (PTC).
Methods
Data from 18,819 patients with PTC from eight university hospitals between January 2012 and February 2018 were retrospectively evaluated. Pathology reports of all PTCs and slides of potential NIFTP cases were reviewed. The strict criterion of no papillae was applied for the diagnosis of NIFTP. Due to assumptions regarding misclassification of NIFTP as non-PTC tumors, the lower boundary of NIFTP prevalence among PTCs was estimated. Mutational analysis for BRAF and three RAS isoforms was performed in 27 randomly selected NIFTP cases.
Results
The prevalence of NIFTP was 1.3% (238/18,819) of all PTCs when the same histologic criteria were applied for NIFTP regardless of the tumor size but decreased to 0.8% (152/18,819) when tumors ≥1 cm in size were included. The mean follow-up was 37.7 months and no patient with NIFTP had evidence of lymph node metastasis, distant metastasis, or disease recurrence during the follow-up period. A difference in prevalence of NIFTP before and after NIFTP introduction was not observed. BRAFV600E mutation was not found in NIFTP. The mutation rate for the three RAS genes was 55.6% (15/27).
Conclusions
The low prevalence and indolent clinical outcome of NIFTP in Korea was confirmed using the largest number of cases to date. The introduction of NIFTP may have a small overall impact in Korean practice.
The follicular variant of papillary thyroid carcinoma (FVPTC) is the second most common type of papillary thyroid carcinoma (PTC) and accounts for 5%–41% of all PTC subtypes [1-4]. FVPTC comprises two major subtypes: infiltrative FVPTC and encapsulated FVPTC (EFVPTC) [5]. The incidence of EFVPTC has gradually increased over the past two decades and constitutes more than 10% of all thyroid malignances diagnosed in North America and Europe [6]. EFVPTC can be further classified into two subtypes: invasive EFVPTC and non-invasive EFVPTC based on the status of capsular or vascular tumor invasion [7]. The majority of non-invasive EFVPTCs behave as indolent non-malignant tumors and have been renamed as non-invasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP) [8]. This introduction of NIFTP concept has significantly affected clinical practice by reducing the therapeutic intensity for patients with EFVPTCs.
Differentiating NIFTP from invasive EFVPTC solely based on cytomorphologic findings is impossible when using thyroid fine needle aspiration cytology. NIFTPs are mostly interpreted as indeterminate categories [9]. The two most common genetic mutations found in thyroid tumors are BRAF and RAS point mutations [10]. RAS mutations occur in 30%–40% of EFVPTCs [11-14]. The BRAFV600E mutation is absent in NIFTPs but can occur in up to 30% of invasive EFVPTCs [14,15].
In Korea, a low rate of NIFTP has been reported in several studies [16-18]. However, the studies were limited by small sample size and single institution, and the results might not represent the general population. Therefore, in the present study, the prevalence of NIFTP among Korean patients was evaluated by reviewing multicenter-based data using the largest cohort size to date in Korea.
MATERIALS AND METHODS
Study cohort
Data from 18,819 patients with PTC from eight university hospitals were retrospectively analyzed. The tumors were diagnosed from surgically resected specimens between January 2012 and February 2018. The slides of all cases initially diagnosed as EFVPTC on pathology reports were reviewed and reclassified into NIFTP, invasive EFVPTC, and other PTC subtypes. The NIFTP was diagnosed according to the criteria of World Health Organization (WHO) Classification of Tumours of Endocrine [19] and recently revised diagnostic criteria [20]: (1) encapsulation or clear demarcation of the tumors (thick, thin, or partial capsule, or well circumscribed with a clear demarcation from adjacent thyroid tissues), (2) follicular growth pattern with no papillae (including microfollicular, normofollicular, or macrofollicular architecture with abundant colloid) with no psammoma bodies and <30% solid/trabecular/insular growth pattern, (3) nuclear score 2–3, (4) no capsular or vascular invasion (requires adequate microscopic examination of the tumor capsule interface), (5) no tumor necrosis, (6) no high mitotic activity (high mitotic activity defined as at least 3 mitoses per 10 high-power fields (400×) (Fig. 1). A nuclear score of 2–3 is diagnostic of NIFTP. However, if florid nuclear features (nuclear score 3) of PTC are present, a meticulous histopathologic examination of the entire tumor is required for the detection of any true papillae, psammoma bodies, aggressive histology, or invasion into tumor capsule or vessels.
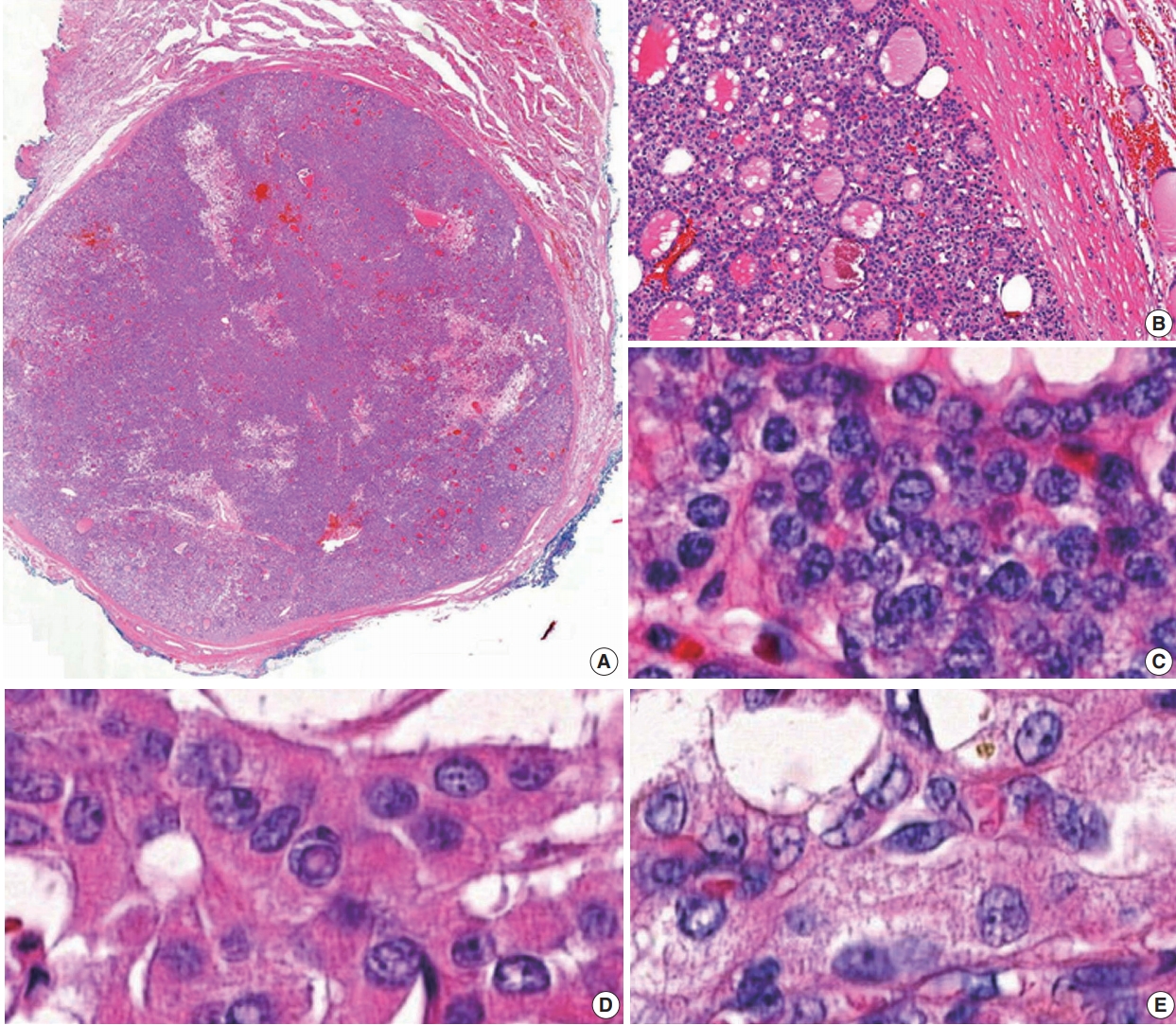
(A) A case of non-invasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP): low power view, NIFTP composed entirely of a follicular growth pattern with complete encapsulation (scan view). (B) NIFTP shows a thick fibrous capsule without capsular and vascular invasion. (C) NIFTP with nuclear score 2. (D, E) Papillary thyroid carcinoma showing nuclear score 3.
Molecular analysis of NRAS and HRAS genes
Molecular analysis of 27 NIFTP cases obtained from a single institution was performed. Genomic DNA was extracted from paraffin-embedded thyroid specimen blocks. The representative slides were selected and the tumor tissues were manually dissected under a stereomicroscope and stored in a 1.5 mL tube. Genomic DNAs were extracted from 5–10-μm-thick tissue sections using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Exon 3 of NRAS and HRAS genes was amplified using polymerase chain reaction (PCR) with the following primers: (1) NRAS-exon 3–197 bp, forward (5'-CCCCTTACCCTCCACACC-3') and reverse (5'-GAGGTTAATATCCGCAAATGACTT-3'); (2) HRAS-exon 3–201 bp, forward (5'-GTCCTCCTGCAGGATTCCTA-3') and reverse (5'-CGGGGTTCACCTGTACT-3'). The PCR cycling conditions for NRAS and HRAS mutations were as follows: initial activation at 94°C for 15 minutes; 35 cycles at 94°C for 30 seconds, 51°C–57°C for 30 seconds, 72°C for 30 seconds, final extension at 72°C for 10 minutes. The amplicons were analyzed using 2% agarose gel electrophoresis and purified using QIAquick PCR purification kit (Qiagen). The amplified PCR products were sequenced using Sanger sequencing. The amplicons were evaluated on 2% agarose gel electrophoresis and purified using the QIAquick PCR purification kit (Qiagen). The amplified PCR products were analyzed using the automated sequencing machine ABI 3730 (Applied Biosystems, Foster City, CA, USA), which performed Sanger sequencing using the same PCR primers [17].
Molecular analysis of KRAS gene
For detection of the KRAS mutation, a PNAClamp KRAS mutation detection kit (Panagene, Daejeon, Korea) was used. The following reagents were used in all PCR reactions (total volume, 20 µL): 10 ng template DNA, primer, peptide nucleic acid (PNA) probe set, and SYBR Green PCR Master Mix. A CFX 96 (Bio-Rad, Hercules, CA, USA) was used for the real-time PCR of PNA-mediated clamping. PCR cycling conditions consisted of the following sequential steps: 5-minute hold at 94°C followed by 40 cycles at 94°C for 30 seconds, 70°C for 20 seconds, 63°C for 30 seconds, and 72°C for 30 seconds. All seven mutations in the KRAS gene were detected using one-step PNA-mediated real-time PCR clamping. PNA probes and DNA primers were used for the corresponding clamping reaction. SYBR Green fluorescent dye was applied for the detection of positive reaction signals. The amplification of the wild-type target was suppressed by the PNA probe sequence complementary to wild-type DNA. Due to this suppression, the amplification of mutant sequences was specifically preferred by competitive inhibition of DNA primers binding to wild-type DNA. The threshold cycle (Ct) value was used to evaluate PCR efficiency. The SYBR Green amplification plots were generated to analyze Ct values for the control and mutation assays. Mutation status was determined by Ct value differences ≥2, which were obtained between the control and samples.
Molecular analysis of BRAF gene
Mutational analysis of BRAF was performed using two different methods. The PNAClamp BRAF mutation detection kit (Panagene) was used to detect the BRAFV600E mutation. Each reaction tube had a total volume of 20 µL and included a mixture of template DNA, primers, PNA probe, and SYBR Green PCR Master Mix. Real-time PCR of PNA-clamping PCR was performed using a CFX96 real-time PCR system (Bio-Rad, Pleasanton, CA, USA). The PNA probe was complementary to wild-type (V600). PCR was performed under the following conditions: 5-minute hold at 94°C, 40 cycles of 30 seconds at 94°C, 20 seconds at 70°C, 30 seconds at 63°C, and 30 seconds at 72°C. The PNA probe and primers incorporated in the assay were separate oligonucleotides, and the PNA probe location was placed between forward and reverse primers within the template. Intercalation of SYBR Green fluorescent dye was used to detect positive signals.
Pyrosequencing for the BRAF mutation analysis was performed as described in detail elsewhere [21]. The primers used for PCR were the following: forward primer (5'-GAAGACCTCACAGTAAAAATAG-3') and reverse primer (5'-biotin-ATAGCCTCAATTCTTACCATCC-3'). The pyrosequencing reaction was performed with a sequencing primer (5'-biotin-ATAGCCTCAATTCTTACCATCC-3') on a Pyromark Q24 instrument (Qiagen). The PyroMark Q24 software (Qiagen) was used for analysis of the pyrogram results.
Statistical analysis
Clinicopathological parameters of NIFTP before and after NIFTP introduction were analyzed using the chi-square test or Fisher’s exact test for categorical variables and the t-test for continuous variables. All statistical analyses were performed using SPSS ver. 22.0 (IBM Corp., Armonk, NY, USA). A p-value of <0.05 was considered statistically significant.
Ethics statement
The current study was approved by the Institutional Review Boards (IRBs) of eight institutions. Ethics approval for all procedures performed in the current study was obtained from the IRB (approval No. 3-2018-0271). Formal written informed consent was waived by the IRB.
RESULTS
The prevalence of NIFTP in PTC cases
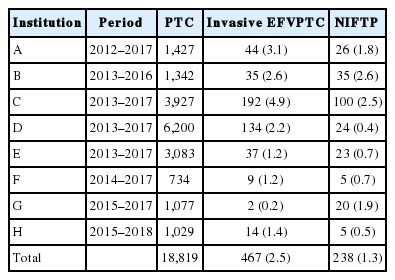
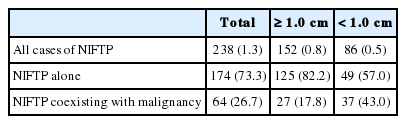
To evaluate the incidence of NIFTP in the Korean population, data were retrospectively collected from the eight university hospitals in Korea. The incidence of NIFTP in each institution is shown in Table 1. After reviewing pathology reports of 18,819 patients with PTC, 378 patients (2.0%) were initially diagnosed with non-invasive EFVPTC as potential NIFTP cases; 140 cases were excluded from NIFTP diagnosis after review of pathology slides acquired from 378 cases. The most common reason for exclusion was the presence of small, but true papillae that were rediagnosed as conventional PTC. Other reasons for exclusion were reclassification as infiltrative FVPTC or unavailability for slide review. Finally, 238 (1.3%) of all PTCs were eligible for the diagnosis of NIFTP after slide review.
Among 238 cases of NIFTP, 174 cases (73.3%) had only NIFTP; in the remaining 64 cases (26.7%), NIFTP coexisted with other malignancies, such as conventional PTC, infiltrative FVPTC, tall cell variant PTC, oncocytic variant PTC, follicular carcinoma, or poorly differentiated carcinoma (Table 2).
Prevalence of NIFTP according to the tumor size among 18,819 patients with initial diagnosis of papillary thyroid carcinoma
Because not all cases were available for slide review and NIFTP can be misclassified as non-PTC tumors, the lower boundary for the prevalence of NIFTP among PTCs was estimated. When the same histologic criteria was applied for the diagnosis of NIFTP regardless of the tumor size, the prevalence of NIFTP was 238 (1.3%) among all PTCs. Among the reclassified 238 NIFTP cases, 86 had a tumor <1.0 cm and 152 had a tumor ≥1.0 cm (Table 2). Therefore, the expected lower boundary for the prevalence of NIFTP was 0.8% (152/18,819) when tumors ≥1.0 cm were included for the diagnosis of NIFTP.
Clinicopathologic characteristics of patients with NIFTP
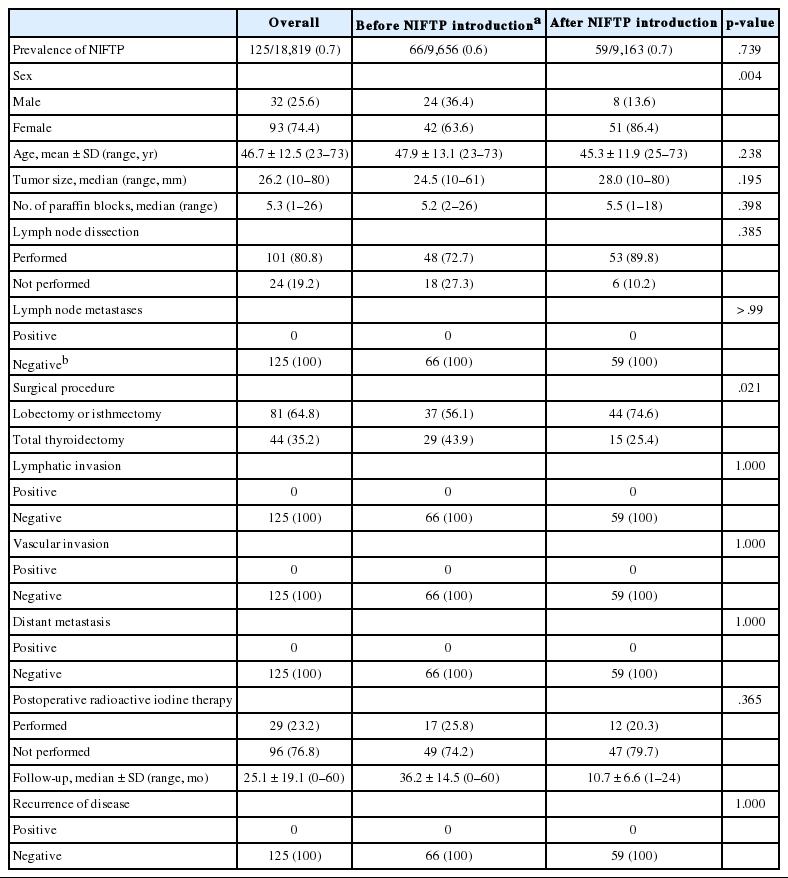
Among 152 patients with NIFTP ≥1.0 cm in size, 125 (82.2%) patients had only NIFTP and the remaining 27 patients (17.8%) had NIFTP coexisting with thyroid cancer. The clinicopathologic characteristics were evaluated in patients with only NIFTP. The demographics of 125 NIFTP patients are shown in Table 3. The mean age was 46.7 years (range, 23 to 73 years). The NIFTP patients included 93 females (74.4%) and 32 males (25.6%). The median primary tumor size was 26.2 mm (range, 10 to 80 mm). Among 125 patients, 101 (80.8%) underwent cervical lymph node dissection and lymph node metastasis was not found. In addition, no patient had lymphatic/vascular invasion or distant metastases. Regarding surgical methods, a total of 44 patients (35.2%) underwent total thyroidectomy and 81 patients (64.8%) received lobectomy or isthmectomy. Furthermore, 29 patients (23.2%) underwent radioactive iodine (RAI) remnant ablation therapy based on the initial tumor size. Disease recurrence was not observed in any NIFTP patient during a median follow-up period of 25.1 months.
Impact of NIFTP on pathologic examination and clinical practice
A difference in the prevalence of NIFTP before and after NIFTP introduction was not observed (Table 3). The number of paraffin blocks for diagnosis of NIFTP did not increase after the introduction of NIFTP (average 5.2 per tumor before April 2016 and average 5.5 per tumor after April 2016).
The rate of lymph node dissection increased from 72.7% before April 2016 to 89.8% after April 2016. The rate of total thyroidectomy decreased from 43.9% before April 2016 to 25.4% after April 2016. The number of patients undergoing postoperative RAI therapy was significantly reduced from 25.8% before April 2016 to 20.3% after April 2016 (Table 3). RAI treatment was performed due to coexisting thyroid cancers in most cases.
BRAF and RAS mutations in NIFTPs
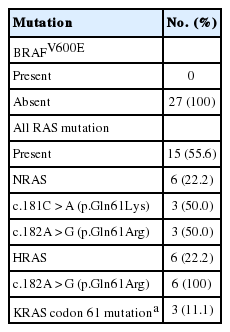
Twenty-seven patients diagnosed with NIFTP at the Gangnam Severance Hospital after April 2016 were randomly selected for gene analysis; BRAFV600E mutation was not observed. As shown in Table 4, the overall frequency of three RAS gene mutations was 55.6% (15/27). The mutation rates of NRAS, HRAS, and KRAS were 22.2%, 22.2%, and 11.1%, respectively.
DISCUSSION
The overall prevalence of NIFTP was 1.3% (range, 0.4% to 2.6%) of all PTCs in the present study, which included the largest cohort size researched to date among the Korean population (Table 1). According to the recent study by Nikiforov et al. [8], any masses <1.0 cm were not included in the diagnosis of NIFTP. Moreover, in another recent NIFTP study, data on the sub-centimeter NIFTP were limited [22]. Thus, the expected lower boundary for the prevalence of NIFTP was 0.8% when tumors ≥1 cm in size were included in the diagnosis of NIFTP.
These results are consistent with the findings from a previous study in which the mean prevalence of NIFTP was 1.5% (range, 0% to 4.7%) in nine institutions from six Asian countries, including Korea [23]. The prevalence of NIFTP is constantly lower in Asian studies than in Western population-based studies [22,24-27].
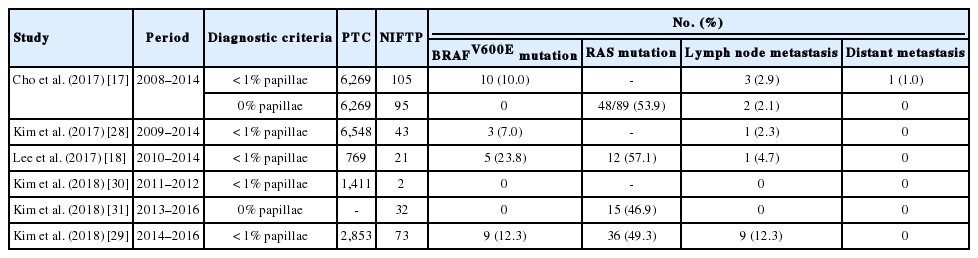
NIFTPs frequently have RAS mutations but no BRAFV600E mutation [11-14]. However, the BRAFV600E mutation was found in some NIFTP cases in several studies immediately performed after the initial publication of NIFTP [17,18,28,29]. Table 5 summarizes the results of BRAF mutation in NIFTP reported in previous studies from six Korean institutions [17,18,28-31]. When the strict criterion of “0% papillae” was applied in the present study, no BRAF mutation was found in NIFTPs. In the present case series, a case of BRAFV600E-positive tumor originally diagnosed as non-invasive EFVPTC was found. The pathology slides were re-examined after cutting deeper sections and a true papillary structure was found in a focal area. Therefore, the case was reclassified as encapsulated classic PTC with predominant follicular growth pattern. This finding reconfirms the strict diagnostic criteria for NIFTP are helpful for excluding true PTC. Because the diagnostic criteria for NIFTP have been updated, NIFTP should no longer include any follicular patterned tumors with well-formed papillae or high-risk gene mutations such as BRAFV600E, TERT promoter, or TP53 [20].
The present study had several technical limitations. First, due to the retrospective multicenter-based nature of this study, interobserver variability in the diagnosis of NIFTP based on histology was not considered. Second, since only the cases diagnosed with PTC were included, the possibility that NIFTP was diagnosed as benign, such as nodular hyperplasia or follicular adenoma, may result in differences in actual prevalence. However, the prevalence of NIFTP did not change significantly even after the introduction of NIFTP. Third, although the overall study cohort size was relatively large, genetic analysis of BRAF and RAS mutations were performed in only a limited number of NIFTP cases. Nevertheless, the results were consistent with previous study results showing NIFTP has no BRAFV600E mutation and frequent RAS mutations [8,15,20].
The results from this study has shown the low prevalence of NIFTP among PTCs in Korea using the largest number of cases to date. Adverse outcomes were not experienced by any patient with NIFTP during the follow-up period. The introduction of NIFTP may have a small overall impact in Korean practice.
Notes
Author contributions
Conceptualization: SWH, CKJ.
Data curation: JYS, JHP, JYP, YJC, CKJ.
Formal analysis: JYS, JHP, JYP.
Funding acquisition: SWH.
Investigation: JYS, JHP, JYP, YJC, CKJ, DES, JJK, SYP, HYN, JHK, JYS, HSK, SWH.
Methodology: JYS, JYP, YJC, CKJ, SWH.
Project administration: CKJ, SWH.
Resources: JYS, JYP, YJC, CKJ, DES, JJK, SYP, HYN, JHK, JYS, HSK, SWH.
Supervision: CKJ, SWH.
Validation: JYS, JYP, YJC, CKJ, SWH.
Visualization: JYS.
Writing—original draft: JYS, CKJ, SWH.
Writing—review & editing: JYS, CKJ, SWH.
Conflicts of Interest
CKJ and SYP, editors-in-chief of the Journal of Pathology and Translational Medicine and SWH, an editorial board member of the Journal of Pathology and Translational Medicine, were not involved in the editorial evaluation or decision to publish this article. All remaining authors have declared no conflicts of interest.
Funding
This research was supported and funded by the Korean Society of Pathologists (grant number: 2017-05-001).