TNF-α and TNF-β Polymorphisms are Associated with Susceptibility to Osteoarthritis in a Korean Population
Article information
Abstract
Background
The tumor necrosis factor (TNF) is believed to play an important role in the pathophysiology of osteoarthritis (OA). Evidence shows that genetic polymorphisms make substantial contributions to the etiology of OA.
Methods
We investigated the genotypes TNF-α and TNF-β in 301 OA patients and 291 healthy subjects as controls. We employed a polymerase chain reaction-restriction fragment length polymorphism and a polymerase chain reaction-single strand conformation polymorphism assay to identify the genotypes TNFA -G308A and TNFB +G252A, respectively.
Results
For TNFA -G308A, the percentages of genotypes GG, AG, and AA were 26.3% (79/301), 62.5% (188/301), and 11.3% (34/301) in OA patients and 88.7% (258/291), 11.3% (33/291), and 0% (0/291) in controls. For TNFB +G252A, the percentages of genotypes GG, AG, and AA were 15.3% (46/301), 41.9% (126/301), and 42.9% (129/301) in OA patients and 12% (35/291), 52.6% (153/291), and 35.4% (103/291) in controls. There were significant differences in genotypes and alleles of TNFA -308 between OA patients and controls (p<0.0001) and in alleles of TNFB +252 (p=0.0325). The risk of OA was significantly higher for carriers of the TNFA -308A allele and the TNFB +252 AA homozygote (p=0.0224).
Conclusions
The results suggest close relationships between TNFA -G308A and TNFB +G252A polymorphisms and individual susceptibility to OA in the Korean population.
Osteoarthritis (OA) is a slowly progressive degenerative joint disease of the articular cartilage that generally occurs in weight-bearing joints and fingers of elderly individuals. OA is characterized by the destruction of the articular cartilage, subchondral bone alterations, and synovitis. Clinical manifestations of OA may include joint pain, swelling, stiffness, and even the loss of some bodily function. The development of OA is a multifactorial process associated with a variety of risk factors, including genetic predispositions, aging, obesity, inflammation, and excessive mechanical loading. In Korea, radiographic and symptomatic knee OA affects 37.3% and 24.2% of elderly individuals, respectively.1 Given the high morbidity and huge economic and personal burdens of OA, there is an urgent need for defining the pathogenesis that contributes to OA development. Recently, new single-nucleotide polymorphisms (SNPs) in the human leukocyte antigen (HLA) class II/III region have been associated with susceptibility to knee OA,2,3 implicating immunologic and inflammatory mechanisms in the etiology and pathophysiology of OA. Both the tumor necrosis factor (TNF)-α and TNF-β genes are located on chromosome 6p21.3 and are closely linked to the HLA.4 This close genetic linkage indicates the possible involvement of the TNF in inflammatory autoimmune diseases.4,5 In addition, polymorphic major histocompatibility complex ancestral haplotypes influence TNF activity.6 Thus, the activity of TNF-α and TNF-β genes may play an important role in the development of knee OA in the Korean population.
The TNF is known as a multifunctional pro-inflammatory cytokine that is involved not only in various physiological processes but also in pathological processes, including inflammation, immunoregulation, proliferation, and apoptosis. TNF-α (also known as cachectin) and TNF-β (formerly known as lymphotoxin) are members of the TNF superfamily. Some of the biological properties of TNF-α and TNF-β suggest that these cytokines may be involved in the destruction of cartilages.7 TNF-α levels are elevated in OA patients' synovial fluid, synovial membrane, subchondral bone, and cartilage.8 However, TNF-β levels are detected differently in inflammatory and autoimmune diseases.
SNPs include some differences or variations in the genome between individuals. Some of these polymorphic variants are functionally expressed, suggesting that SNPs are among the factors associated with susceptibility to diseases. The transcriptional regulation of the TNF gene is essential for avoiding the deleterious effects of the inappropriate or excessive synthesis of the TNF.9 Therefore, genetic variations within the TNF promoter may influence the transcription and expression of the TNF. Polymorphisms in the promoter region of the TNF gene have been detected at -G238A, -G308A, -C863A, and -C857A in the TNF-α promoter and at +G252A and +G318C in intron 1 of the TNF-β gene. The up-regulation of TNF-α expression is involved in the pathogenesis of a large variety of illnesses with inflammatory and autoimmune components. Previous research has demonstrated that TNF-α production is higher in carriers of the -308A allele than in -308G carriers, indicating that this polymorphism has functional implications for transcriptional activation and subsequent increases in inflammation.10 The TNFB +252G allele has been associated with higher TNF-β production at both the mRNA and protein levels.11,12 However, some studies have found that the TNFB +252A allele is more likely to increase TNF-α secretory capacity and plasma TNF-α levels than the TNFB +252G allele.13 In view of these considerations, this study focuses on the genetic susceptibility of TNF-α and TNF-β polymorphisms for OA.
To determine the relationships between TNF-α and TNF-β gene polymorphisms and individual susceptibility to OA in a Korean population, this study examines the percentages of genotypes and alleles for the TNF-α polymorphism (TNFA -G308A) by the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and the TNF-β polymorphism (TNFB +G252A) by the polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) in 301 patients with OA and 291 healthy individuals.
MATERIALS AND METHODS
Tissue samples
We obtained degenerative articular cartilage, meniscus, and ligament tissue specimens from 301 OA patients who received total knee arthroplasty at St. Mary's Hospital of The Catholic University of Korea between 2004 and 2005. All patients were confirmed by radiology and pathology to have OA. Among the 301 patients, 51 (16.9%) were male, and 250 (83.1%) were female. The mean age was 60 at the initial diagnosis. Because these patients received total knee arthroplasty, we obtained the specimens from those patients of Kellgren-Lawrence grade 4 or joint space narrowing grade 4 or higher. We excluded those patients with rheumatoid arthritis (RA), polyarthritis-associated autoimmune diseases, post-traumatic OA, and infection-induced OA. We also excluded those with clinical and radiographic findings suggesting skeletal dysplasia and those with other malignant diseases such as bone tumors, secondary metastasis, alcohol/drug dependence, hepatic failure, and renal failure. The control group consisted of a total of 291 healthy individuals (130 females and 161 males). The mean age was 51. We excluded those individuals with joint pain, a limp, or limited joint movement and those with radiographic signs of joint space narrowing or osteophyte formation. The healthy individuals and patients belonged to the same ethnicity and geographical area. This study was approved by the Institutional Review Board (IRB) of the Catholic University of Korea, College of Medicine (IRB approval No. CUMC10U177).
DNA extraction
We extracted DNA templates from the paraffin-embedded knee joint tissue of OA patients. We cut these tissue samples into 4-5 µm slices and dissolved the paraffin with xylene, then washed the xylene with 100% ethanol. We suspended the tissues in an ice-cold Nonidet P-40 lysis buffer solution and treated them with proteinase K. We extracted the DNA by the phenol/chloroform/isoamyl alcohol extraction method and ethanol precipitation, following previous research.14 For healthy individuals, we obtained a leukocyte cell pellet from each blood sample through the Buffy coat by the centrifugation of 2 mL of whole blood. We used the cell pellet for the DNA extraction. We employed the Qiagen DNA blood mini kit (Qiagen, Valencia, CA, USA) and followed the manufacturer's instructions to obtain the genomic DNA. We determined the purity and concentration of the extracted DNA by using the Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA).
PCR-RFLP for TNF-α
We used a PCR-RFLP assay to identify the TNFA -G308A genotype. The primers were sense 5'-AGG CAA TAG GTT TTG AGG GCC AT-3' and antisense 5'-TCC TCC CTG CTC CGA TTC CG-3'. We performed each PCR procedure under standard conditions with a 10 µL PCR mixture containing 1 µL of template DNA, 0.5 µM of each primer, 0.2 µM of each deoxynucleotide triphosphate, 1.5 mM of MgCl2, 0.4 U of the AmpliTaq gold polymerase (Perkin-Elmer, Foster City, CA, USA), and 1 µL of 10× buffer. We denatured the reaction mixture for 12 minutes at 94℃ and incubated it for 40 cycles (denaturation for 30 seconds at 94℃, annealing for 30 seconds at 60℃, and elongation for 30 seconds at 72℃). We continued the final extension for 5 minutes at 72℃. After the amplification, we digested the PCR products with 5 U of the restriction enzyme NCO1 for 4 hours at 37℃. We then separated the digested product on a 3% agarose gel containing ethidium bromide and photographed it by using the Ultra Violet Product Image Store system (Ultraviolet Products, Upland, CA, USA) (Fig. 1A). To ensure the reliability of the RFLP results, we sequenced the PCR products by using the fluorescent dideoxy chain termination method with the ABI 3730XL Analyzer (Applied Biosystems, Foster City, CA, USA) based on the manufacturer's instructions (Fig. 1B).
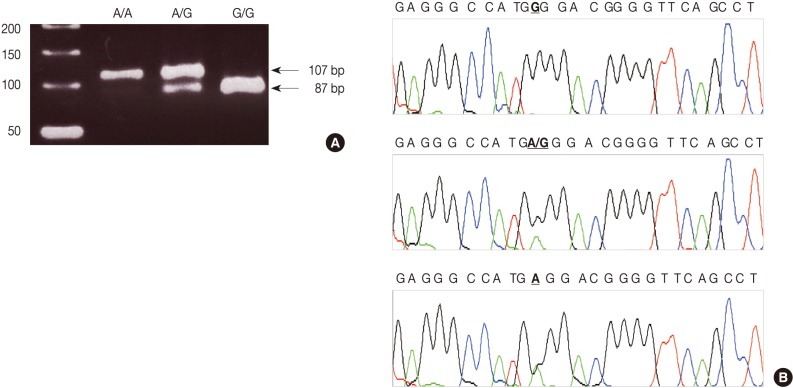
Genotypes of the tumor necrosis factor (TNF)-α polymorphism by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). (A) The NCO1 restriction enzyme recognize and cut the PCR product of the G-type allele into 2 fragments. Lane 1, A/A; lane 2, A/G; lane 3, G/G; lane M, 50-base pair ladder. (B) Sequencing analysis shows three genotypes at the -G308A site of the TNF-α gene: homozygotes with G/G (upper panel) and A/A (bottom panel) genotypes and a heterozygote with the A/G (middle panel) genotype.
PCR-SSCP for TNF-β
We employed a PCR-SSCP assay to identify the TNFB +G252A genotype. The primers were sense 5'-CTC CTG CAC CTG CTG CCT GGA TC-3' and antisense 5'-GAA GAG ACG TTC AGG TGG TGT CAT-3'. We performed each PCR procedure under standard conditions with a 10 µL PCR mixture containing 1 µL of template DNA, 0.5 µM of each primer, 0.2 µM of each deoxynucleotide triphosphate, 1.5 mM of MgCl2, 0.4 U of the AmpliTaq gold polymerase (Perkin-Elmer), 0.5 µCi of [32P]dCTP (Amersham, Buckinghamshire, UK), and 1 µL of 10× buffer. We denatured the reaction mixture for 12 minutes at 95℃ and incubated it for 40 cycles (denaturation for 30 seconds at 95℃, annealing for 30 seconds at 60℃, and elongation for 30 seconds at 72℃). We continued the final extension for 5 minutes at 72℃. After the amplification, we denatured the PCR products for 5 minutes at 95℃ in a 1:1 dilution of a sample buffer solution containing 98% formamide and 5 mmol/L NaOH. We loaded these products onto an SSCP gel (FMC Mutation Detection Enhancement system, Intermountain Scientific, Kaysville, UT, USA) containing 10% glycerol. After the electrophoresis, we transferred the gels to 3 MM Whatman paper and dried them. We then performed the autoradiography by using Kodak X-OMAT film (Eastman Kodak, Rochester, NY, USA) (Fig. 2A). We cut the DNA showing mobility shifts from the dried gels and amplified it for 40 cycles by using the same primer set. We sequenced the PCR products by using the ABI 3730XL Analyzer (Applied Biosystems) (Fig. 2B). Blinded to the status of the study cohort, one of the authors evaluated the results. We randomly selected more than 10% of the sample for repeated assays and found that the results were in complete agreement.
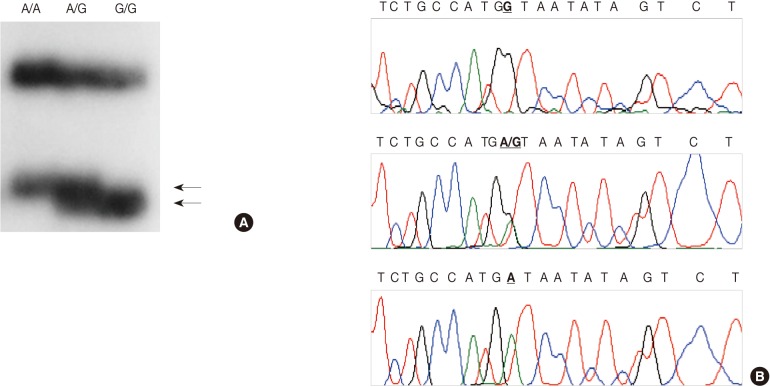
Genotypes of the tumor necrosis factor (TNF)-β polymorphism by polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP). (A) SSCP demonstrating bands of each genotype. Lane 1, A/A; lane 2, A/G; lane 3, G/G. (B) Sequencing analysis shows three genotypes at the +G252A site of the TNF-β gene: homozygotes with G/G (upper panel) and A/A (bottom panel) genotypes and a heterozygote with the A/G (middle panel) genotype.
Statistical analysis
We conducted a two-tailed Fisher's exact test to determine the differences in the percentages of genotypes and alleles between OA patients and controls. Genotype-specific risk was estimated as an odds ratio at the 95% confidence interval, when we conducted a multiple logistic regression analysis to estimate the interaction between TNF-α and TNF-β. Because both the TNF-α and TNF-β genes are located on chromosome 6p21.3, and the TNF-β polymorphism is associated with TNF-α secretory capacity and the plasma TNF-α level,4,9,10 we analyzed the relationships between the assembled haplotypes and the risk of OA by conducting a logistic regression analysis.
RESULTS
TNFA -G308A polymorphism and the risk of OA
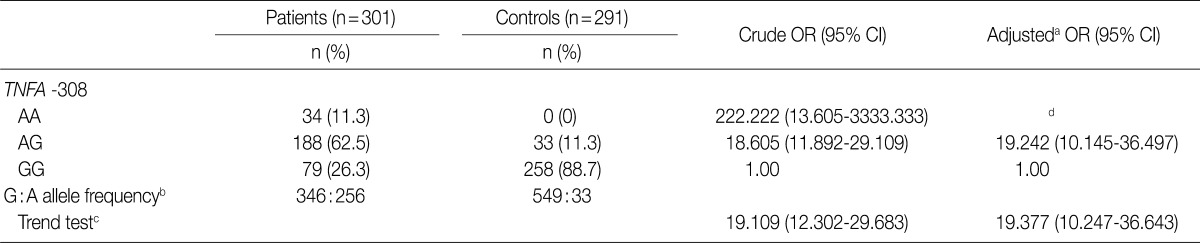
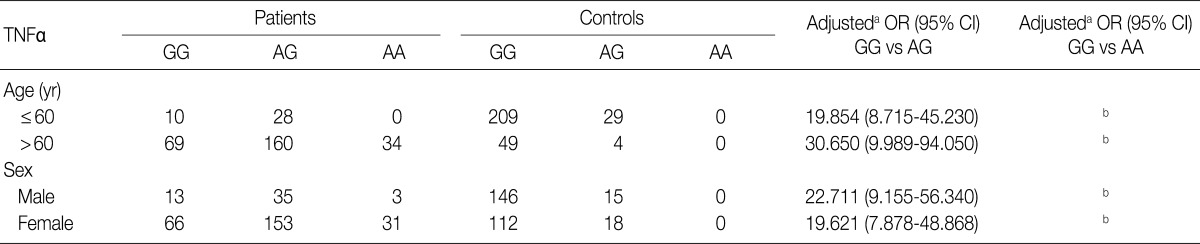
We considered 301 OA patients and 291 healthy individuals (as controls). Table 1 shows the distributions of genotypes and alleles for the TNFA -G308A polymorphism in OA patients and controls. For healthy individuals, the percentages of genotypes GG, AG, and AA were 88.7% (258/291), 11.3% (33/291), and 0% (0/291), respectively, and the percentages of alleles G and A were 94.3% and 5.7%, respectively. Unexpectedly, healthy individuals did not carry the A/A homozygosity of TNFA -G308A. For OA patients, the percentages of genotypes GG, AG, and AA were 26.3% (79/301), 62.5% (188/301), and 11.3% (34/301), respectively, and the percentages of alleles G and A were 57.5% and 42.5%, respectively. There were significant differences in genotype and allele frequencies for TNFA -308 between OA patients and controls (p<0.0001). We could not estimate the relationships between TNFA -G308A polymorphism genotypes and OA stratified according to age and sex (Table 2).
TNFB +G252A polymorphism and the risk of OA
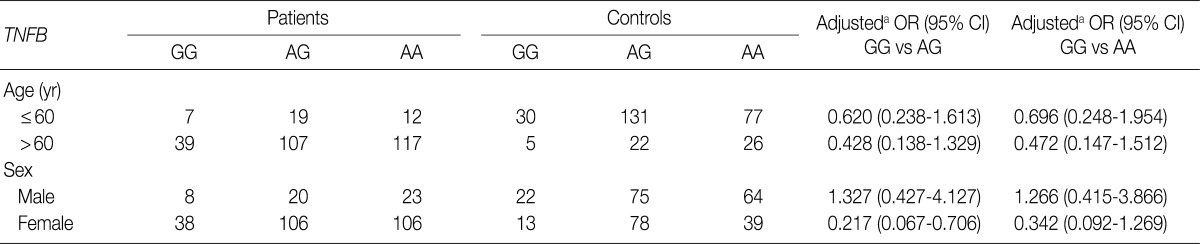
Table 3 shows the distributions of genotypes and alleles for the TNFB +G252A polymorphism in OA patients and controls. For controls, the percentages of genotypes GG, AG, and AA were 12% (35/291), 52.6% (153/291), and 35.4% (103/291), respectively, and the percentages of alleles G and A were 38.3% and 61.7%, respectively. For OA patients, the percentages of genotypes GG, AG, and AA were 15.3% (46/301), 41.9% (126/301), and 42.9% (129/301), respectively, and the percentages of alleles G and A were 36.2% and 63.8%, respectively. There were significant differences in the percentages of alleles for TNFB +252 between OA patients and controls (p=0.0325). However, there was no significant difference in the genotypic distribution of TNFB +252 (p=0.4542). Table 4 shows the relationship between the TNFB +G252A polymorphism genotype and OA stratified according to age and sex. Because the typical age for OA onset among Koreans is approximately 60, we classified the patients into two age groups: ≤60 as "young" patients and >60 as "old" patients. As shown in Table 4, there was no significant difference between OA patents stratified according to age and sex.
Haplotypes TNF-α and TNF-β and the risk of OA
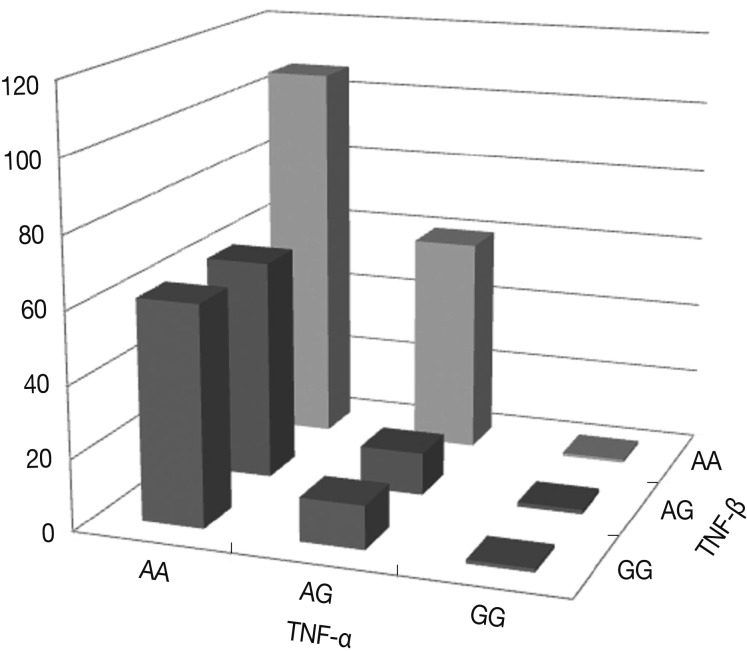
Fig. 3 shows the percentages of genotypes and alleles for the assembled haplotypes TNFA -G308A and TNFB +G252A in OA patients. The results of the logistic regression analysis for the relationships between the assembled haplotypes and the risk of OA indicate that all genotypes with the A allele of the TNF-α gene carried more than a tenfold increase in the risk of OA compared with those genotypes with the G allele of both the TNF-α and TNF-β genes (p<0.0001). There was a significant interaction in terms of the percentages of the assembled haplotypes between TNFA -G308A and TNFB +G252A. In particular, the genotypes with the AA allele of both the TNF-α and TNF-β genes carried more than a hundred fold increase in the risk of OA compared with those genotypes with the GG allele. The risk of OA was significantly higher for carriers of the TNFA -308A allele and the TNFB +252 AA homozygote (p=0.0224).
DISCUSSION
Recent studies have shown that some genetic susceptibility contribute to the etiology of OA, including EDG2, ASPN, CALM1, COL2A1, COMP, and FRZB.15 Over the years, it has become evident that the inflammatory cytokine network contributes substantially to the pathogenesis of OA. An increase in the expression of pro-inflammatory cytokines in the patient's cartilage, synovial membrane, and subchondral bone is believed to be linked to the development and progression of structural changes in the OA joint.15 A number of studies have provided evidence of the participation of the TNF family in OA.15 Genetic variations within the TNF promoter may influence the transcription and expression of the TNF. In addition, the up-regulation of TNF gene expression is known to be involved in the pathogenesis of a large variety of illnesses with inflammatory and autoimmune components.
TNF-α appears to play a pivotal role in the imbalance between anabolic and catabolic processes of OA patients. It can down-regulate the synthesis of major extracellular matrix (ECM) components through the inhibition of the anabolic activity of chondrocytes.16 For instance, TNF-α suppresses the synthesis of link proteins and type II collagen, which are major components of the ECM.17 The TNF also stimulates chondrocytes to release matrix metalloproteinases (MMPs), which have the capacity to degrade cartilage matrix proteins.18 Transgenic mice expressing high concentrations of TNF-α develop arthritis spontaneously.9 SNPs of TNF-α have been found at -863, -857, -308, and -238 sites in the promoter.19 Among the polymorphisms described within the TNF-α promoter, TNFA -G308A has been found to be linked to RA, systemic lupus erythematosus (SLE), and ankylosing spondylitis.13,20 The less common A allele is strongly associated with elevated TNF levels and disease susceptibility in human subjects.21 In this study, the percentages of genotypes at the -G308A site of the TNF-α gene in OA patients were 26.3% for GG, 62.5% for AG, and 11.3% for AA. There was a significant difference in the percentage of the -G308A polymorphism of TNF-α between OA patients and controls (p<0.0001) (Table 1). In addition, there was a significant difference in the allele percentage between OA patients and controls (p<0.0001) (Table 1). These results indicate that the TNFA -308A allele may be a genetic predisposing factor in susceptibility to OA in the Korean population.
TNF-β is predominantly expressed in B- and T-lymphocytes and natural killer cells. However, its expression has also been described in chondrocytes.22 It has been implicated in lymphoid follicle development and the production of pro-inflammatory cytokines and can facilitate the proliferation of fibroblasts and synoviocytes.23 TNF-β is the homologue of TNF-α, and both bind to TNF-RI and TNF-RII receptors and share many common biological activities. Even at low levels, TNF-β is more likely to induce the secretion of interleukin (IL)-6, IL-8, and MMP3 than TNF-α.24-26 TNF-β is an important pro-inflammatory mediator and biomarker in RA, SLE, and sepsis.24-28 However, the levels of TNF-β production are detected differently in these inflammatory and autoimmune diseases. An important polymorphism influencing TNF-β expression has been found in intron 1 of the gene at the nucleotide position +A252G, and the polymorphism is associated with the risk of RA, SLE, sepsis, pancreatic cancer, and breast cancer.27,28 A high level of the TNFB +252G allele has been found in patients with pancreatic cancer and breast cancer.29 TNFB +252A is associated with the increased risk of developing RA, SLE, and sepsis.24-28 In this study, the percentages of genotypes at the +G252A site of the TNF-β gene in OA patients were 15.3% for GG, 41.9% for AG, and 42.9% for AA. In terms of genotype percentages, there was no significant difference between OA patients and controls (p=0.4542) (Table 3). However, there was a significant difference in the allele percentage at the +G252A site of the TNF-β gene between OA patients and controls (p=0.0325) (Table 3). This suggests that although the A allele of TNFB +A252G is associated with lower TNF-β production,11,12,30 the A allele can increase TNF-α secretory capacity. Thus, the A allele genotype may be a genetic predisposing factor in susceptibility to OA in the Korean population. In addition, there was a significant interaction between TNF-α and TNF-β genes and the occurrence of OA. This suggests that the A allele of the TNF-α gene is the susceptibility gene of OA and that the risk of OA is significantly higher in carriers of the TNFA -308A allele and the TNFB +252 AA homozygote (p=0.0224).
In conclusion, we demonstrated the relationships of the -G30 8A polymorphism in the TNF-α gene promoter and the +G25 2A polymorphism in intron1 of the TNF-β gene with susceptibility to OA of the knee in a Korean population. Future molecular genetic research should consider larger samples to determine the genetic susceptibility of the TNF-α and TNF-β polymorphisms and identify the mechanisms underlying these polymorphisms.
Acknowledgments
This research was supported by the Public Welfare & Safety Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology (2010-0020764).
Notes
No potential conflict of interest relevant to this article was reported.