Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 56(2); 2022 > Article
-
Case Study
An unusual case of microsatellite instability–high/deficient mismatch repair (MSI-H/dMMR) diffuse large B-cell lymphoma revealed by targeted gene sequencing -
Bogyeong Han1
 , Sehui Kim1
, Sehui Kim1 , Jiwon Koh1
, Jiwon Koh1 , Jeong Mo Bae1
, Jeong Mo Bae1 , Hongseok Yun2
, Hongseok Yun2 , Yoon Kyung Jeon1,3
, Yoon Kyung Jeon1,3
-
Journal of Pathology and Translational Medicine 2022;56(2):92-96.
DOI: https://doi.org/10.4132/jptm.2021.10.15
Published online: November 16, 2021
1Department of Pathology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
2Center for Precision Medicine, Seoul National University Hospital, Seoul, Korea
3Cancer Research Institute, Seoul National University, Seoul, Korea
- Corresponding Author: Yoon Kyung Jeon, MD, PhD, Department of Pathology, Seoul National University Hospital, Seoul National University College of Medicine, 103 Daehak-ro, Jongno-gu, Seoul 03080, Korea, Tel: +82-2-740-8323, Fax: +82-2-743-5530, E-mail: ykjeon@snu.ac.kr
© 2022 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (https://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
- Microsatellite instability-high/deficient mismatch repair (MSI-H/dMMR) status has been approved as a tissue-agnostic biomarker for immune checkpoint inhibitor therapy in patients with solid tumors. We report the case of an MSI-H/dMMR diffuse large B-cell lymphoma (DLBCL) identified by targeted gene sequencing (TGS). A 90-year-old female who presented with vaginal bleeding and a large mass in the upper vagina was diagnosed with germinal center-B-cell-like DLBCL, which recurred at the uterine cervix at 9 months after chemotherapy. Based on TGS of 121 lymphoma-related genes and the LymphGen algorithm, the tumor was classified genetically as DLBCL of EZB subtype. Mutations in multiple genes, including frequent frameshift mutations, were detected by TGS and further suggested MSI. The MSI-H/dMMR and loss of MLH1 and PMS2 expression were determined in MSI-fragment analysis, MSI real-time polymerase chain reaction, and immunohistochemical tests. This case demonstrates the potential diagnostic and therapeutic utility of lymphoma panel sequencing for DLBCL with MSI-H/dMMR.
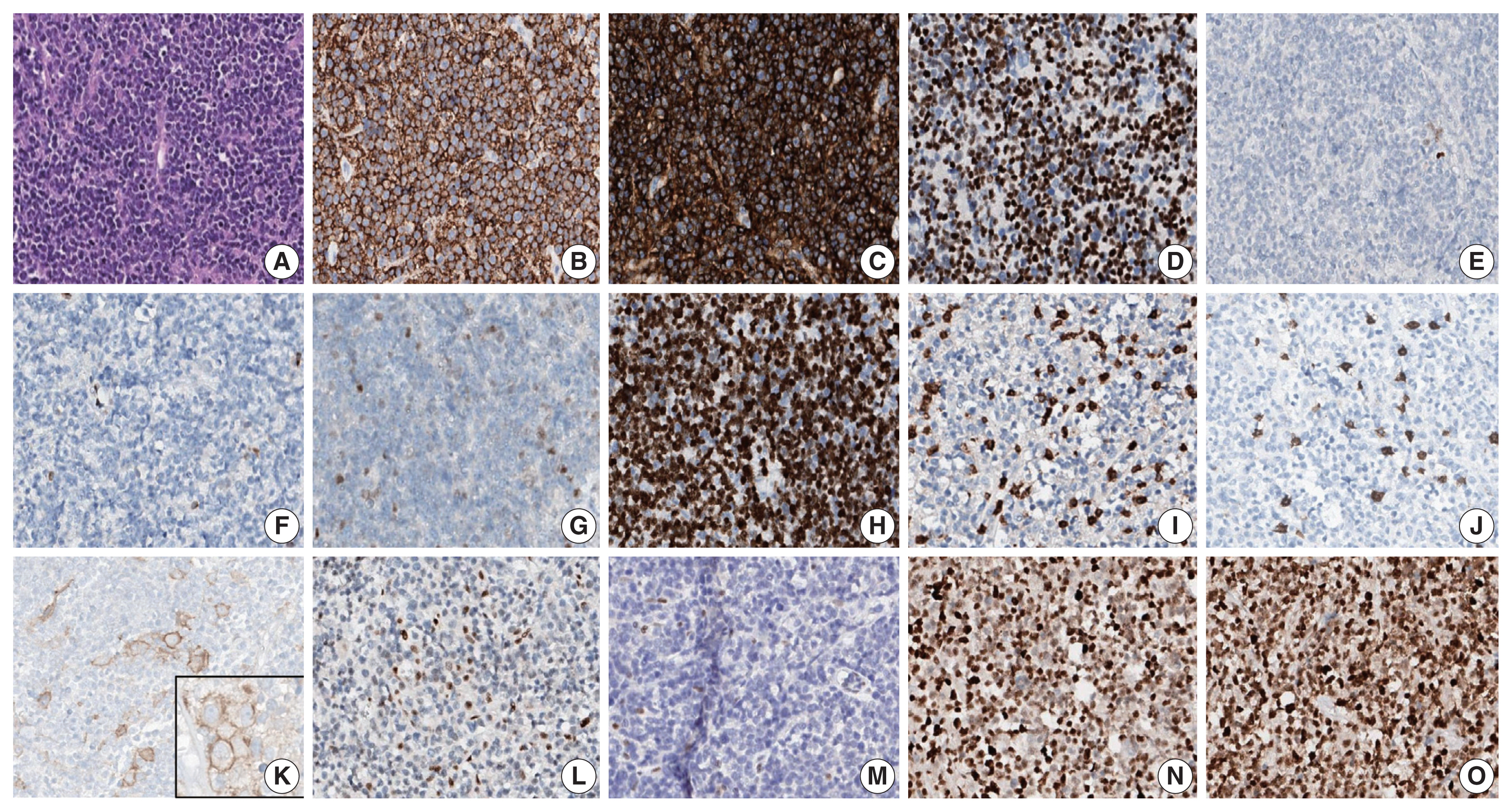
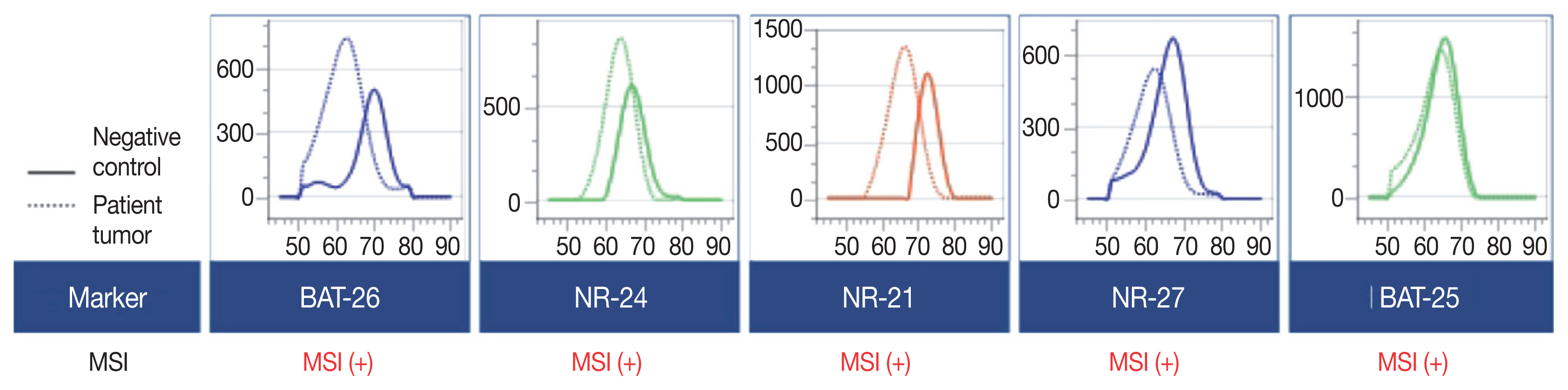
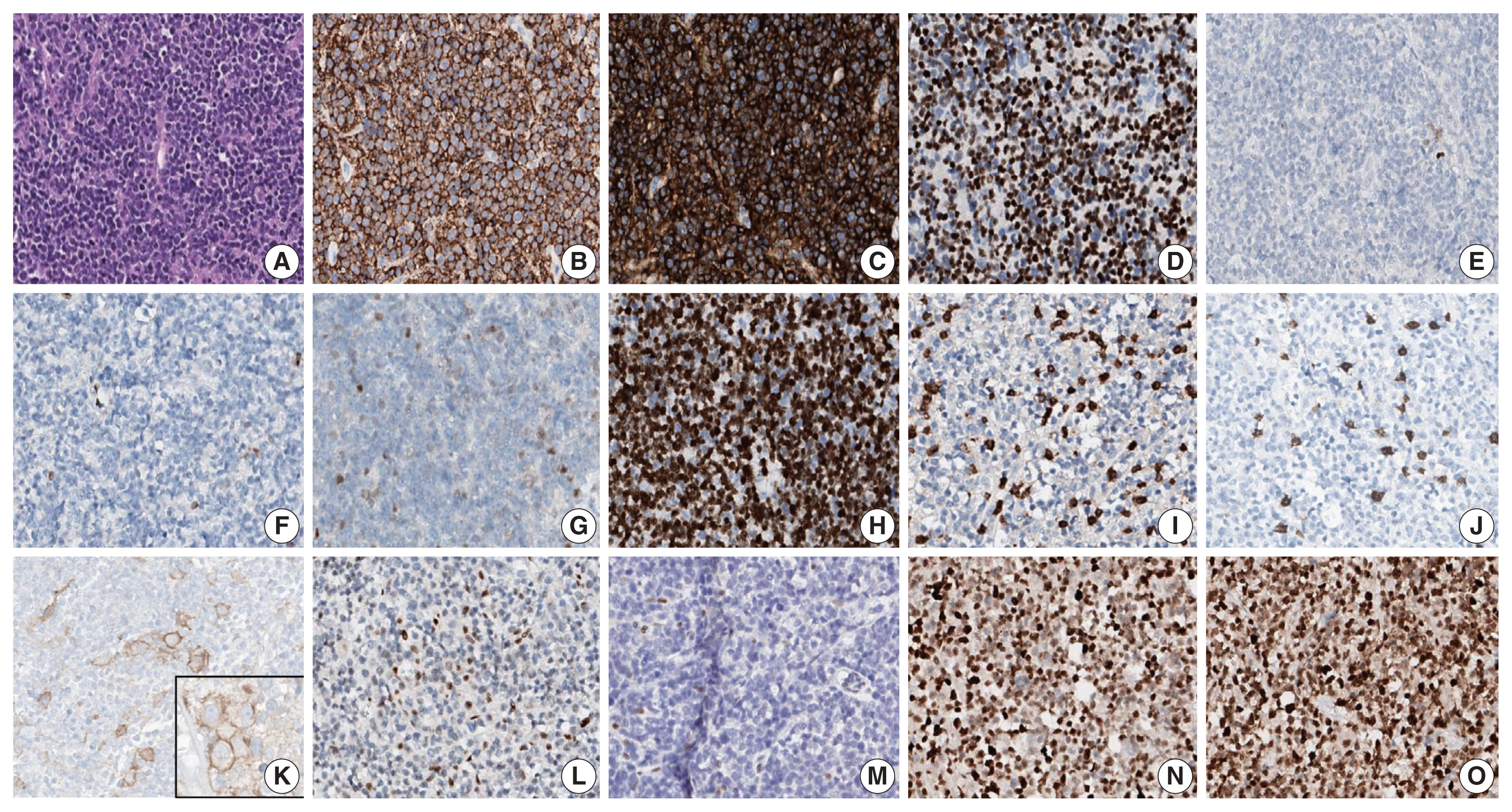
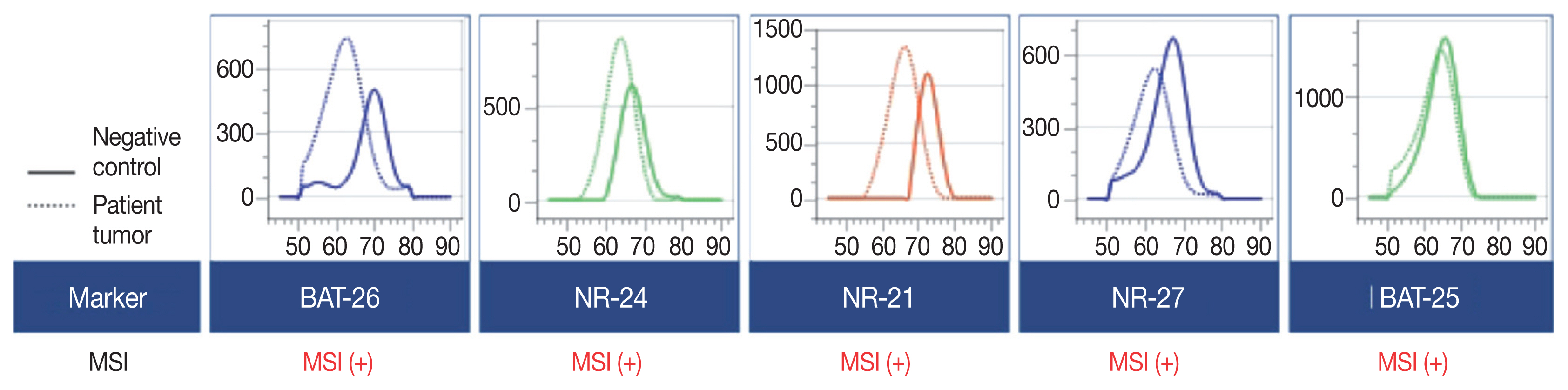
- A 90-year-old female presented with vaginal bleeding. A protruding polypoid mass measuring 5.8 cm was detected in the upper vagina by pelvic magnetic resonance imaging. Positron emission tomography revealed an additional focal hypermetabolic lesion in the presacral area. A tissue biopsy was conducted under the suspicion of cervical cancer. The patient was diagnosed with DLBCL with a germinal center B-cell-like (GCB) phenotype determined by immunohistochemistry (IHC)-based Hans algorithm, clinical stage of IIA, and Eastern Cooperative Oncology Group performance status of 1. Rituximab and reduced-dose CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone) resulted in complete remission (CR), but after 9 months, she suffered local relapse at the uterine cervix and underwent tumor excision. Microscopically, the tumor was composed of atypical lymphoid cells with a centroblastic morphology and diffuse arrangement. IHC for CD3, CD20, C-MYC, BCL-2, BCL-6, CD10, MUM1, Ki-67, CD8, and programmed death-ligand 1 (PD-L1) (clone E1L3N) and in situ hybridization for Epstein-Barr virus (EBV) and fluorescence in situ hybridization (FISH) for MYC were performed. Tumor cells were positive for CD20, CD10, and BCL-6 but negative for MUM1, BCL-2, C-MYC, and EBV. The Ki-67 labeling index of tumor cells was 80% (Fig. 1A–H). MYC translocation was not observed in FISH (data not shown). The tumor was diagnosed again as DLBCL with a GCB phenotype. Scattered CD3+ or CD8+ tumor-infiltrating lymphocytes were observed (Fig. 1I, J), and PD-L1 was expressed in tumor-associated macrophages and in about 5% of tumor cells (Fig. 1K). TGS was performed using a customized panel comprising 121 lymphoma-related genes and formalin-fixed paraffin-embedded sections of the surgical specimen; it revealed 16 mutations across 15 genes including ARID1A, DNMT3A, PDCD1, SETD2, PDGFRA, TET2, PRDM1, CARD11, ATM, KMT2D, B2M, CREBBP, CIITA, GNA13, and BTK (Table 1). The tumor was genetically classified as EZB subtype DLBCL according to the LymphGen algorithm (https://llmpp.nih.gov/lymphgen/index.php) [10]. In addition, 34 further mutations (27 missense mutations, 5 frameshift mutations, 1 inframe indel mutation, and 1 stop-gain mutation) in 28 genes were identified as rare germline variants or variants of uncertain significance (data not shown). Although microsatellite markers were not included in this lymphoma panel, the unusually high number of variants, including a large number of frameshift mutations, suggested MSI. A fragmentation assay based on the Bethesda guidelines, immunohistochemistry of four proteins involved in the mismatch repair (MMR) pathway (MLH1, MSH2, MSH6 and PMS2), and a U-TOP MSI detection test (Seasun Biomaterials Inc., Daejeon, Korea) were conducted. On the fragmentation assay, four (BAT25, BAT26, D2S123, and D17S2720) of the five Bethesda microsatellite markers showed features suggestive of MSI (data not shown). A loss of MLH1 and PMS2 protein expression was detected by immunohistochemistry (Fig. 1L–O), and the U-TOP MSI detection test, which uses five quasi-monomorphic mononucleotide markers that do not require samples of normal tissue from the patient for comparison [11], showed MSI in all five markers, confirming the MSI-H/dMMR status of the tumor (Fig. 2). The patient was treated by radiotherapy of the involved site and has been in metabolic CR for the past 7 months.
CASE REPORT
- MSI-H or dMMR status is the first tissue-agnostic biomarker approved by the FDA for cancer therapy and, more specifically, for pembrolizumab therapy [3]. Patients with MSI-H/dMMR tumors have shown a favorable clinical response to PD-1 blockade, regardless of cancer type [2]. This illustrates the need for MSI-H tumor detection. MSI-PCR of the Bethesda panel to assess three dinucleotide repeats (D2S123, D5S346, D175250) and two mononucleotide repeats (BAT26, BAT25) is the gold standard for detecting MSI. According to the revised Bethesda guidelines, pentaplex PCR with five quasi-monomorphic mononucleotide repeats can detect MSI with high sensitivity and specificity and might not need matched normal tissue [1]. Together with immunohistochemistry for MMR proteins, it allows determination of MMR status [1]. MSI testing performed in endometrial, colorectal, and gastric cancers has revealed high variability in the frequency of MSI-H among cancers [4,12]. However, the frequency of MSI-H non-Hodgkin lymphoma generally is low (0%–2%), which has hampered standardized MSI testing as a routine diagnostic method [4,12]. Recently, next-generation sequencing (NGS)-based TGS using a panel of tumor markers has been introduced for molecular pathologic diagnosis. In solid tumors, NGS-based MSI tests have shown high sensitivity and specificity in the absence of control normal tissue [4–7]. Although unified criteria for detection of MSI by NGS are lacking, many laboratories are using MMR-related genes and MSI markers, as well as bioinformatics algorithms, for tumor diagnosis [4–7].
- The FDA has approved PD-1 blockade therapy based on its efficacy in patients with hematologic malignancies, including Hodgkin lymphoma and primary mediastinal large B-cell lymphoma [13,14]. A recent study demonstrated the potential benefit of PD-1 blockade in combination with R-CHOP in patients with treatment-naive DLBCL overexpressing PD-L1 [15]. However, both the MSI landscape and efficacy of PD-l blockade for MSI-H lymphoma remain unclear. A previous study reported DLBCL with defects in DNA repair genes, including tumor suppressor genes, MMR-related genes (e.g., MSH2 and MSH6), and non-homologous end-joining pathway-related genes, and it suggested an association of genomic instability phenotype with tumorigenesis of DLBCL [16]. However, recent analyses of NGS data showed that MSI-H in non-Hodgkin lymphoma, including DLBCL, is either extremely rare (<1%) or not a feature of these tumors [4–7]. In contrast to those reports, a study using a commercial MSI kit, based on eight mononucleotide repeat markers and two pentanucleotide repeat markers, identified MSI-H and microsatellite instability–low (MSI-L) in 3% (3/92) and 10% (9/92) of DLBCLs, respectively [9]. According to the authors, patients with MSI-H DLBCL tended to have a better prognosis than those with microsatellite stable DLBCL, although the difference was not significant [9]. They also reported that MSI-L DLBCL was associated with a poor response to chemotherapy [9]. These results suggest that MSI status could be a useful biomarker for DLBCL.
- Reports on the prognostic and therapeutic impacts of molecular genetic classification of DLBCL indicate the potential utility of TGS in individually tailored treatment for DLBCL [10,17]. The LymphGen algorithm classified DLBCLs into five genetic subtypes: MCD (including MYD88 L265P and CD79B mutations), BN2 (including BCL6 translocations and NOTCH2 mutations), N1 (including NOTCH1 mutations), EZB (including EZH2 mutations and BCL2 translocations), and A53 (aneuploid with TP53 inactivation) [10]. Our inclusion of 121 lymphoma-related genes allowed successful genetic determination of the tumor as EZB, which is the most common genetic subtype of GCB-DLBCL [10]. TGS also revealed frequent frameshift mutations and C:G → A:T transversions, as previously reported in MMR-mutated DLBCLs [16]. Although MMR gene mutations could not be identified directly using our lymphoma panel, the unique mutational pattern of our patient’s tumor was suggestive of an MSI-H/dMMR DLBCL. The MSI status of the tumor was confirmed by two kinds of MSI tests for microsatellite markers (i.e., MSI-fragmentation assay and MSI-pentaplex real-time PCR using U-TOP MSI detection test), as well as by immunohistochemistry for four MMR proteins.
- However, because our patient did not receive PD-l blockade therapy, the efficacy for treating this type of tumor could not be determined. Nonetheless, this case shows that MSI-H/dMMR can be present in DLBCL, albeit rarely, and the utility of TGS for detection of MSI-H/dMMR in hematolymphoid malignancies.
DISCUSSION
Ethics Statement
This study was approved by the Institutional Review Board (IRB) of Seoul National University Hospital (No. 2012-160-1184) and written informed consent from the patient was waived by IRB decision.
Availability of Data and Material
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
Author Contributions
Conceptualization: YKJ, JMB. Data curation: BH, SK. Formal analysis: JK, JMB, HY. Investigation: BH, JMB, HY. Visualization: BH, SK. Writing—original draft: BH, SK. Writing—review & editing: JK, YKJ. Approval of final manuscript: all authors.
Conflicts of Interest
The authors declare that they have no potential conflicts of interest.
Funding Statement
This study was funded by Basic Science Research Program (grant No.: NRF-2016R1D1A1B01015964) through the National Research Foundation (NRF) funded by the Ministry of Education, Science and Technology (MEST).
- 1. Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 2004; 96: 261-8. PubMed
- 2. Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017; 357: 409-13. PubMedPMC
- 3. Marcus L, Lemery SJ, Keegan P, Pazdur R. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res 2019; 25: 3753-8. ArticlePubMedPDF
- 4. Bonneville R, Krook MA, Kautto EA, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol 2017; 2017: PO.17.00073. Article
- 5. Middha S, Zhang L, Nafa K, et al. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO Precis Oncol 2017; 2017: PO.17.00084. Article
- 6. Albayrak A, Garrido-Castro AC, Giannakis M, et al. Clinical pan-cancer assessment of mismatch repair deficiency using tumor-only, targeted next-generation sequencing. JCO Precis Oncol 2020; 4: 1084-97. ArticlePubMedPMC
- 7. Trabucco SE, Gowen K, Maund SL, et al. A novel next-generation sequencing approach to detecting microsatellite instability and pan-tumor characterization of 1000 microsatellite instability-high cases in 67,000 patient samples. J Mol Diagn 2019; 21: 1053-66. ArticlePubMedPMC
- 8. Duval A, Raphael M, Brennetot C, et al. The mutator pathway is a feature of immunodeficiency-related lymphomas. Proc Natl Acad Sci U S A 2004; 101: 5002-7. ArticlePubMedPMC
- 9. Tian T, Li J, Xue T, Yu B, Li X, Zhou X. Microsatellite instability and its associations with the clinicopathologic characteristics of diffuse large B-cell lymphoma. Cancer Med 2020; 9: 2330-42. ArticlePubMedPMCPDF
- 10. Wright GW, Huang DW, Phelan JD, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell 2020; 37: 551-68. ArticlePubMedPMC
- 11. Suraweera N, Duval A, Reperant M, et al. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology 2002; 123: 1804-11. ArticlePubMed
- 12. Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 2016; 22: 1342-50. ArticlePubMedPDF
- 13. Armand P, Rodig S, Melnichenko V, et al. Pembrolizumab in relapsed or refractory primary mediastinal large B-cell lymphoma. J Clin Oncol 2019; 37: 3291-9. PubMedPMC
- 14. Kuruvilla J, Ramchandren R, Santoro A, et al. Pembrolizumab versus brentuximab vedotin in relapsed or refractory classical Hodgkin lymphoma (KEYNOTE-204): an interim analysis of a multicentre, randomised, open-label, phase 3 study. Lancet Oncol 2021; 22: 512-24. ArticlePubMed
- 15. Smith SD, Till BG, Shadman MS, et al. Pembrolizumab with R-CHOP in previously untreated diffuse large B-cell lymphoma: potential for biomarker driven therapy. Br J Haematol 2020; 189: 1119-26. ArticlePubMedPDF
- 16. de Miranda NF, Peng R, Georgiou K, et al. DNA repair genes are selectively mutated in diffuse large B cell lymphomas. J Exp Med 2013; 210: 1729-42. ArticlePubMedPMCPDF
- 17. Dubois S, Viailly PJ, Mareschal S, et al. Next-generation sequencing in diffuse large B-cell lymphoma highlights molecular divergence and therapeutic opportunities: a LYSA study. Clin Cancer Res 2016; 22: 2919-28. ArticlePubMedPDF
REFERENCES
Figure & Data
References
Citations

- Shared genomic features of HIV+ diffuse large B-cell lymphoma in two African cohorts
Sophia M. Roush, Mishalan Moodley, Jenny Coelho, Samantha Beck, Amon Chirwa, Edwards Kasonkanji, Marriam Mponda, Maurice Mulenga, Tamiwe Tomoka, Hanri van Zijl, Katherine Hodkinson, Arshad Ismail, Senzo Mtshali, Jonathan Featherston, Satish Gopal, Matthew
Scientific Reports.2025;[Epub] CrossRef - Chimeric and mutant CARD9 constructs enable analyses of conserved and diverged autoinhibition mechanisms in the CARD‐CC protein family
Jens Staal, Yasmine Driege, Femke Van Gaever, Jill Steels, Rudi Beyaert
The FEBS Journal.2024; 291(6): 1220. CrossRef - PD-L1+diffuse large B-cell lymphoma with extremely high mutational burden and microsatellite instability due to acquiredPMS2mutation
Andrew W. Allbee, James Gerson, Guang Yang, Adam Bagg
Molecular Case Studies.2023; 9(4): a006318. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure


Fig. 1
Fig. 2
| Gene | Chr | Position | Reference sequence | Exon | cDNA change | AA change | VAF (%) |
|---|---|---|---|---|---|---|---|
| ARID1A | chr1 | 27105930 | NM_006015.4 | 20 | c.5548dupG | p.Asp1850fs | 34.32 |
| DNMT3A | chr2 | 25457242 | NM_022552.4 | 23 | c.2645G > A | p.Arg882His | 44.45 |
| PDCD1 | chr2 | 242795103 | NM_005018.2 | 2 | c.105delC | p.Thr36fs | 36.23 |
| SETD2 | chr3 | 47125614 | NM_014159.6 | 12 | c.5656G > A | p.Glu1886Lys | 41.73 |
| PDGFRA | chr4 | 55151558 | NM_006206.4 | 17 | c.2347delT | p.Ser783fs | 42.52 |
| TET2 | chr4 | 106193857 | NM_001127208.2 | 10 | c.4319G > A | p.Arg1440Gln | 44.39 |
| PRDM1 | chr6 | 106555015 | NM_001198.3 | 7 | c.2132C > A | p.Ala711Asp | 43.55 |
| CARD11 | chr7 | 2983911 | NM_032415.5 | 5 | c.619C > T | p.Arg207Cys | 42.73 |
| ATM | chr11 | 108216612 | NM_000051.3 | 58 | c.8561G > A | p.Arg2854His | 40.91 |
| KMT2D | chr12 | 49420238 | NM_003482.3 | 48 | c.15511C > T | p.Arg5171Trp | 43.12 |
| KMT2D | chr12 | 49431873 | NM_003482.3 | 34 | c.9265dupG | p.Val3089fs | 36.08 |
| B2M | chr15 | 45003779 | NM_004048.2 | 1 | c.35T > C | p.Leu12Pro | 44.60 |
| CREBBP | chr16 | 3786070 | NM_004380.2 | 28 | c.4694delA | p.Lys1565fs | 42.97 |
| CIITA | chr16 | 11001304 | NM_001286402.1 | 11 | c.1965dupC | p.Gly656fs | 82.74 |
| GNA13 | chr17 | 63052509 | NM_006572.5 | 1 | c.203T > G | p.Met68Arg | 43.85 |
| BTK | chrX | 100613407 | NM_000061.2 | 12 | c.993A > G | p.Ile331Met | 45.38 |
Chr, chromosome; AA, amino acid; VAF, variant allelic frequency.

 E-submission
E-submission