Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 59(5); 2025 > Article
-
Original Article
Unraveling the crucial role of CCL3 in nasopharyngeal carcinoma: bioinformatics and immunohistochemical insights -
Xiaopeng Guo,*
 , Zhen Sun,*
, Zhen Sun,* , Ya Liang
, Ya Liang , Aoshuang Chang
, Aoshuang Chang , Junjun Ling
, Junjun Ling , Houyu Zhao
, Houyu Zhao , Xianlu Zhuo
, Xianlu Zhuo
-
Journal of Pathology and Translational Medicine 2025;59(5):281-290.
DOI: https://doi.org/10.4132/jptm.2025.05.23
Published online: June 20, 2025
Department of Otorhinolaryngology Head and Neck Surgery, Affiliated Hospital of Guizhou Medical University, Guiyang, China
-
Corresponding Author: Xianlu Zhuo, MD Department of Otorhinolaryngology Head and Neck Surgery, Affiliated Hospital of Guizhou Medical University, Guiyang, Guizhou 550004, China Tel: +86-851-86855119, Fax: +86-851-86855119, E-mail: zhuoxianlu@gmc.edu.cn
Houyu Zhao, MD Department of Otorhinolaryngology Head and Neck Surgery, Affiliated Hospital of Guizhou Medical University, Guiyang, Guizhou 550004, China Tel: +86-851-86855119, Fax: +86-851-86855119, E-mail: zhaohouyujia@163.com - *These authors contributed equally to this work.
© The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
- 1,199 Views
- 132 Download
Abstract
-
Background
- C-C motif chemokine ligand 3 (CCL3) is a crucial chemokine that plays a fundamental role in the immune microenvironment and is closely linked to the development of various cancers. Despite its importance, there is limited research regarding the expression and function of CCL3 in nasopharyngeal carcinoma (NPC). Therefore, this study seeks to examine the expression of CCL3 and assess its clinical significance in NPC using bioinformatics analysis and experiments.
-
Methods
- The bioinformatics approach was employed to assess the expression and function of CCL3 in NPC. Subsequently, protein expression of CCL3 was detected in an NPC cohort using immunohistochemistry based on a tissue microarray. The relationship between CCL3 expression and clinical features was then investigated.
-
Results
- A total of 20 CCL3-related genes and 14 possible target genes were identified through bioinformatics analysis, many of which play crucial roles in pathways such as chemokine signaling pathway and transcriptional misregulation in cancer signaling pathways. CCL3 was found to be associated with drug resistance and various immune cell infiltrations. In NPC, CCL3 expression was significantly higher than normal controls, and high expression of CCL3 correlated with cervical lymph node metastasis, tumor recurrence, advanced clinical stage, and poor prognosis.
-
Conclusions
- CCL3 may be a key gene in the initiation and progression of NPC. It has the potential to serve as both a diagnostic biomarker and a therapeutic target for NPC.
- Nasopharyngeal carcinoma (NPC), a malignant neoplasm arising from the epithelial cells of the nasopharynx, is influenced by a confluence of factors including genetics, environmental exposures, and Epstein-Barr virus infection [1]. While the incidence and mortality of NPC have decreased in recent years due to lifestyle changes and advancements in healthcare [2], the disease remains highly prevalent in certain regions like southern China. Unfortunately, there remains a lack of reliable methods for the early diagnosis, effective assessment, and prognostic prediction of NPC [3]. Current treatment modalities for NPC predominantly involve radiotherapy and chemotherapy [1]. Although patients diagnosed at an early stage often benefit from standalone radiotherapy, a significant proportion approximately 75% to 90% are identified at advanced local stages [4]. Additionally, those undergoing radical radiotherapy are at a heightened risk for recurrence and distant metastasis [5]. Local recurrence and distant metastases also pose significant challenges and account for 95% of NPC-related deaths [6]. Moreover, the toxicity associated with radiotherapy can substantially impair the quality of life for patients, regardless of their overall prognosis [5]. Thus, the pursuit of innovative strategies for early diagnosis and novel treatment approaches remains of paramount importance.
- In the process of tumorigenesis, multiple factors are involved, with abnormal expression of proto-oncogenes and dysregulation of oncogenes potentially playing crucial roles. Current research on NPC highlights the significance of specific genes, such as EPHB2 [7], whose elevated expression is associated with poor prognosis, and TRIM21 [8], which may correlate with adverse outcomes and early tumor relapse following radiotherapy. The aberrant expression of these genes could be pivotal in NPC development. Consequently, some researchers are investigating targeted gene therapy approaches for NPC treatment. For instance, combining radiotherapy with programmed death-ligand 1/programmed death-1 checkpoint inhibitors has been shown to improve radiotherapy efficacy against NPC [9]. Similarly, integrating targeted epidermal growth factor receptor therapy with induction chemotherapy and concurrent radiotherapy has proven effective in treating locally advanced NPC [10]. However, these treatment strategies remain in the exploratory phase, and identifying additional early diagnostic markers and therapeutic targets continues to be of critical clinical significance.
- CCL3 (C-C motif chemokine ligand 3 or macrophage inflammatory protein 1 alpha) is an inducible chemokine involved in immune surveillance and tolerance [11]. In the tumor microenvironment, chemokines play pivotal roles in regulating immune cells and shaping the immune landscape [11]. Previous studies have demonstrated that CCL3 enhances colorectal cancer proliferation via the TRAF6 (tumor necrosis factor receptor-associated factor 6)/nuclear factor кB signaling pathway [12]. Moreover, CCL3 has emerged as a potential biomarker for diagnosing oral squamous cell carcinoma [13]. However, there is limited research on the expression of CCL3 in NPC.
- In this study, we employed bioinformatics methods to investigate CCL3 expression in NPC tissues and to further assess its role in NPC.
INTRODUCTION
- Analysis of the GSE53819 dataset
- To investigate the expression of CCL3 in NPC tissues, we selected a dataset from the Gene Expression Omnibus (GEO) database for validation. GSE53819 [14], which includes 18 NPC samples and 18 normal tissue samples, was chosen to analyze the differential expression of CCL3 between NPC tissues and normal tissues.
- Drug susceptibility prediction
- The Genomics of Drug Sensitivity in Cancer (GDSC) database [15] is a public resource that provides information on molecular markers of drug sensitivity and drug response in cancer cells, encompassing nearly 700 cancer cell lines and 138 anticancer drugs. We utilized this tool to predict the potential impact of key gene expression on drug sensitivity and visualized the results.
- Immunological correlation analysis
- To investigate the relationship between the expression of CCL3 and the tumor immune microenvironment, we utilized the Tumor Immune Estimation Resource (TIMER) database [16] to assess the correlation between CCL3 expression and the infiltration of various immune cells. Concurrently, based on the expression matrix of the GSE53819 dataset, we analyzed the immune infiltration abundance in NPC using the Immune Cell Abundance Identifier platform [17], and further evaluated the correlation between CCL3 expression and infiltration levels of diverse immune cell subtypes through the CIBERSORT algorithm combined with Spearman correlation analysis.
- Prediction of intergenic interactions involving CCL3
- The STRING database [18] was used to predict and analyze the protein-protein interaction (PPI) relationship. This database allows the construction of comprehensive PPI networks from a list of genes. To explore the potential biological functions of CCL3, a PPI network involving CCL3 was constructed using the STRING database. Genes closely associated with CCL3 were filtered out, and the PPI network was employed to gain insights into the numerous interactions between genes/proteins. Additionally, the Cytoscape software [19] was utilized to calculate the degree and betweenness centrality of each node in the PPI network and visualize the PPI network. Nodes with higher scores play more critical roles in the network. The NetworkAnalyst tool [20] was employed to predict the target genes of CCL3.
- The genes that may interact with CCL3 and the target genes obtained earlier were imported into the DAVID database [21] to predict their potential roles by using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The results in descending order based on the number of participating genes were sorted. The top-ranked analysis results were visualized separately.
- Immunohistochemistry staining and evaluation of the relationship between CCL3 and clinicopathological features
- Tissue microarray (TMA) has proved to be an effective and efficient tool for assessing quality assurance programs such as intra- and interlaboratory variation in immunohistochemical (IHC) and molecular studies [22]. A total of 129 NPC samples were included in a TMA (HNasN129Su01: number of cores: 129; core diameter: 1.5 mm; section thickness: 4 μm) provided by Shanghai Outdo Biotech Co, Ltd. (Shanghai, China). The cohort consisted of 99 males and 30 females, aged between 20 to 82 years. Diagnosis was performed between January 2010 and October 2011 and the last follow-up took place in March 2017. All cases were diagnosed with NPC through clinicopathological assessments and none received preoperative radiotherapy. This TMA presents clinicopathological characteristics of all cases.
- CCL3 protein expression was evaluated via IHC staining with a CCL3 rabbit polyclonal antibody (1:500, BIOSS, Beijing, China) according to the manufacturer’s guidelines. Negative controls were included by omitting the primary antibody (secondary antibody and chromogen only), and Ki-67 (1:4,000, Ki-67 antibody, Cat# 66555-6-Ig, clone 1B9H2, host species: mouse, Proteintech Group, Inc., Rosemont, IL, USA) served as a positive control to validate experimental procedures. The IHC staining outcomes were assessed by a composite score based on the intensity and percentage of positivity. In summary, the staining intensity was rated from 0 to 3, where 0, 1, 2, and 3 indicated negative, weak, moderate, and strong staining, respectively. The percentage of positive expression was rated on a scale from 0 (0%–5%), 1 (6%–25%), 2 (26%–50%), 3 (51%–75%), to 4 (76%–100%). The overall IHC score was calculated by multiplying the staining intensity and the percentage of positive expression. The IHC score ranged from 0 to 12, where 0 to 6 indicated low expression and >6 denoted high expression [23]. The IHC outcomes were evaluated independently and blindly by two observers who were uninformed of the clinical parameters of the cases. The correlation between CCL3 and clinicopathological features was examined.
- Statistical analysis
- Differences between groups were analyzed using an analysis of variance (ANOVA), a t-test, or a Wilcoxon rank sum test, depending on the specific type of data, using MedCalc software [24]. The chi-square test was used to distinguish the rates of different groups. p < .05 was considered statistically significant.
MATERIALS AND METHODS
Construction of protein-protein interaction networks and prediction of target genes
Exploration of the possible functions of genes that might interact with CCL3
- Expression of CCL3 in NPC tissues
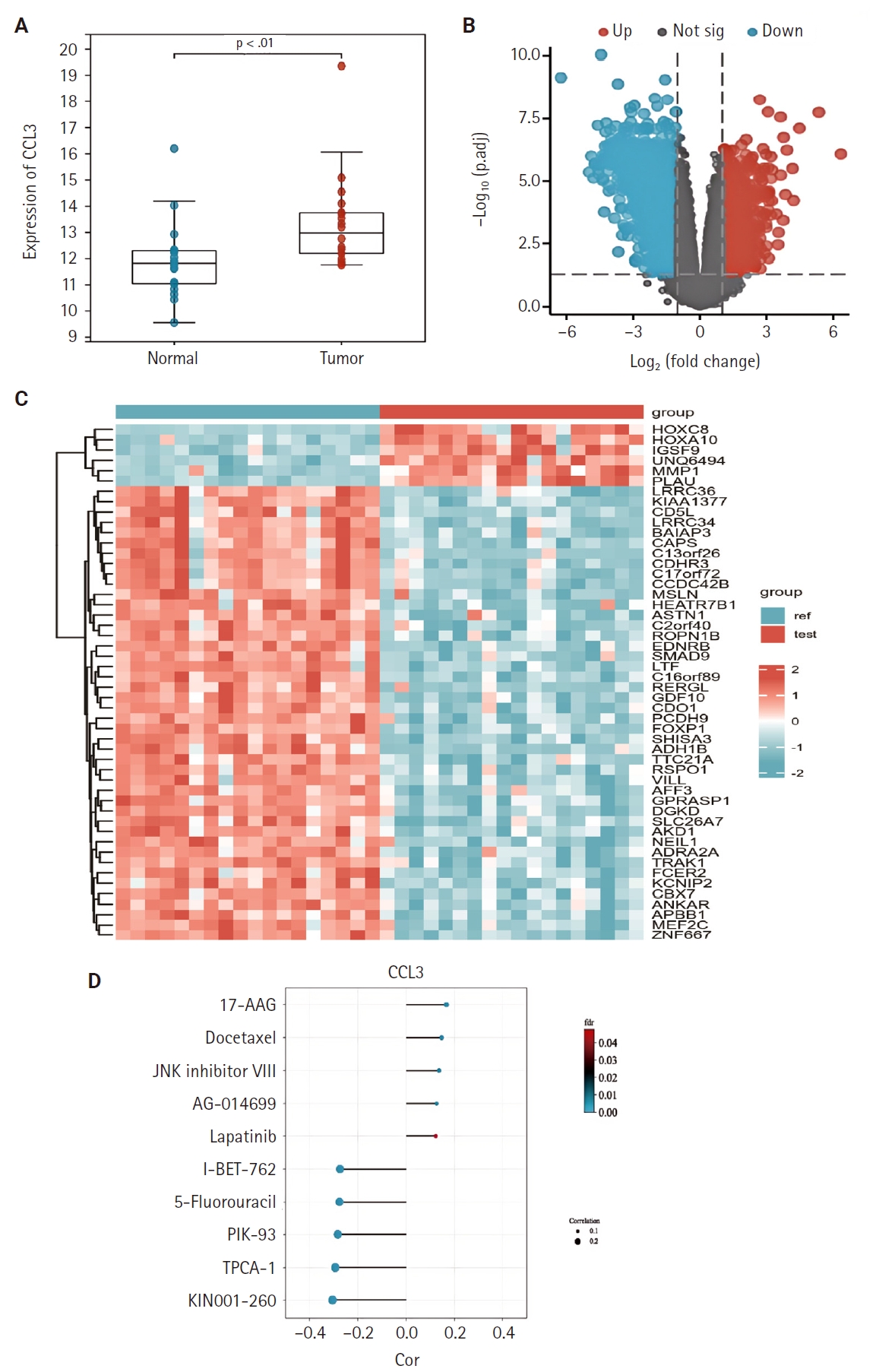
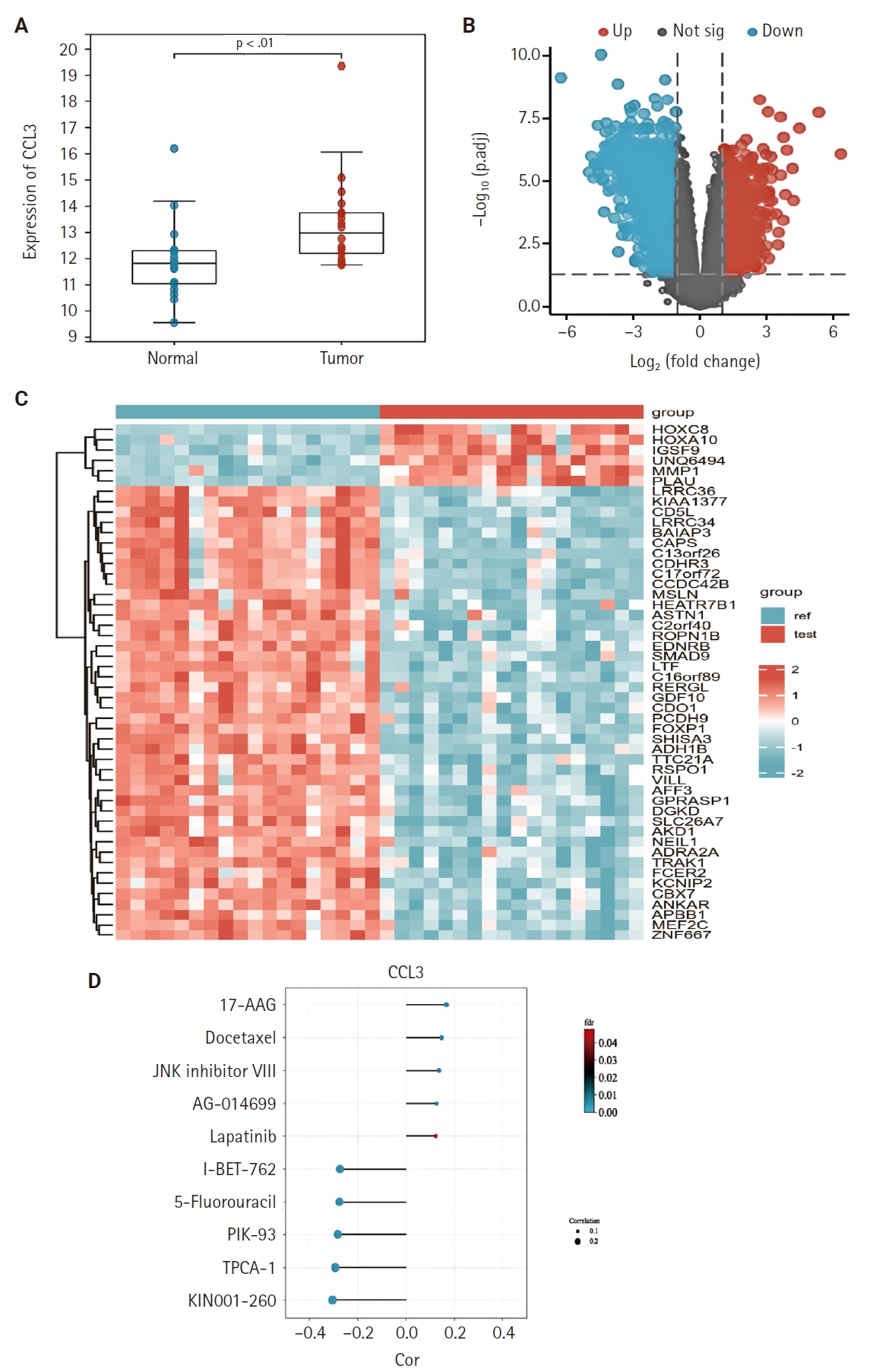
- We examined the differential expression of CCL3 between NPC tissues and normal tissues using the GSE53819. As shown in Fig. 1A, CCL3 exhibited significantly higher expression levels in NPC tissues relative to normal tissues. Additionally, differential analysis of tumor versus normal tissues in the GSE53819 dataset identified a heatmap of genes exhibiting upregulated and downregulated expression in NPC (Fig. 1B, C).
- Correlation of CCL3 expression with the cancer cell sensitivity to multiple chemotherapeutic agents
- The GDSC database was employed to predict the potential impact of CCL3 expression on the chemosensitivity of cancer cells. Our analysis revealed a negative correlation between CCL3 expression and the sensitivity of cancer cells to various drugs. For visualization purposes, we selected several drugs with a strong correlation, including I-BET-762, 5-fluorouracil, PIK-93, TPCA-1, and KIN001-260 (Fig. 1D), Notably, 5-fluorouracil, a commonly used chemotherapeutic agent for NPC, may have its efficacy modulated by CCL3 expression in NPC.
- Correlation analysis between CCL3 gene expression and immune cell infiltration
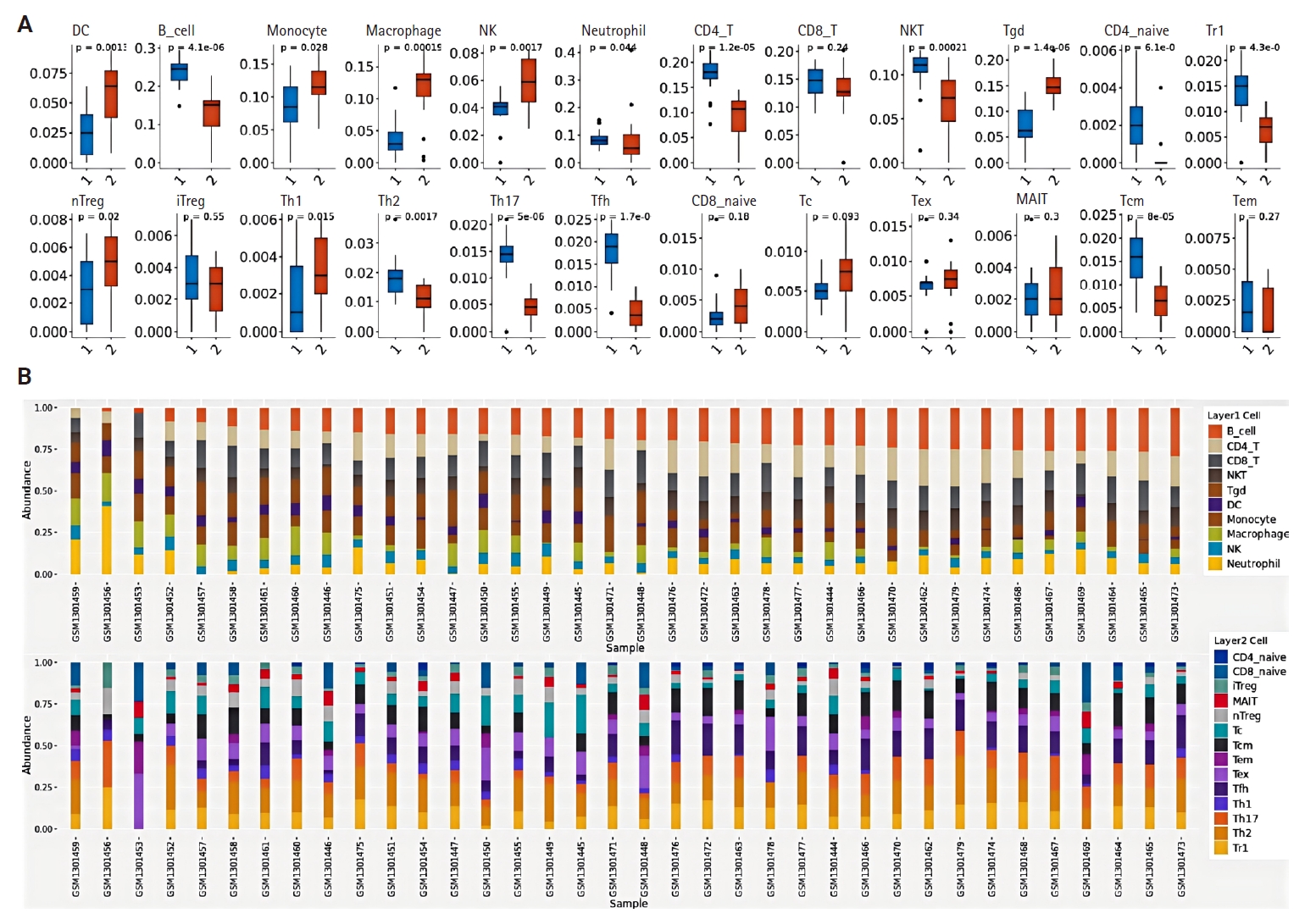
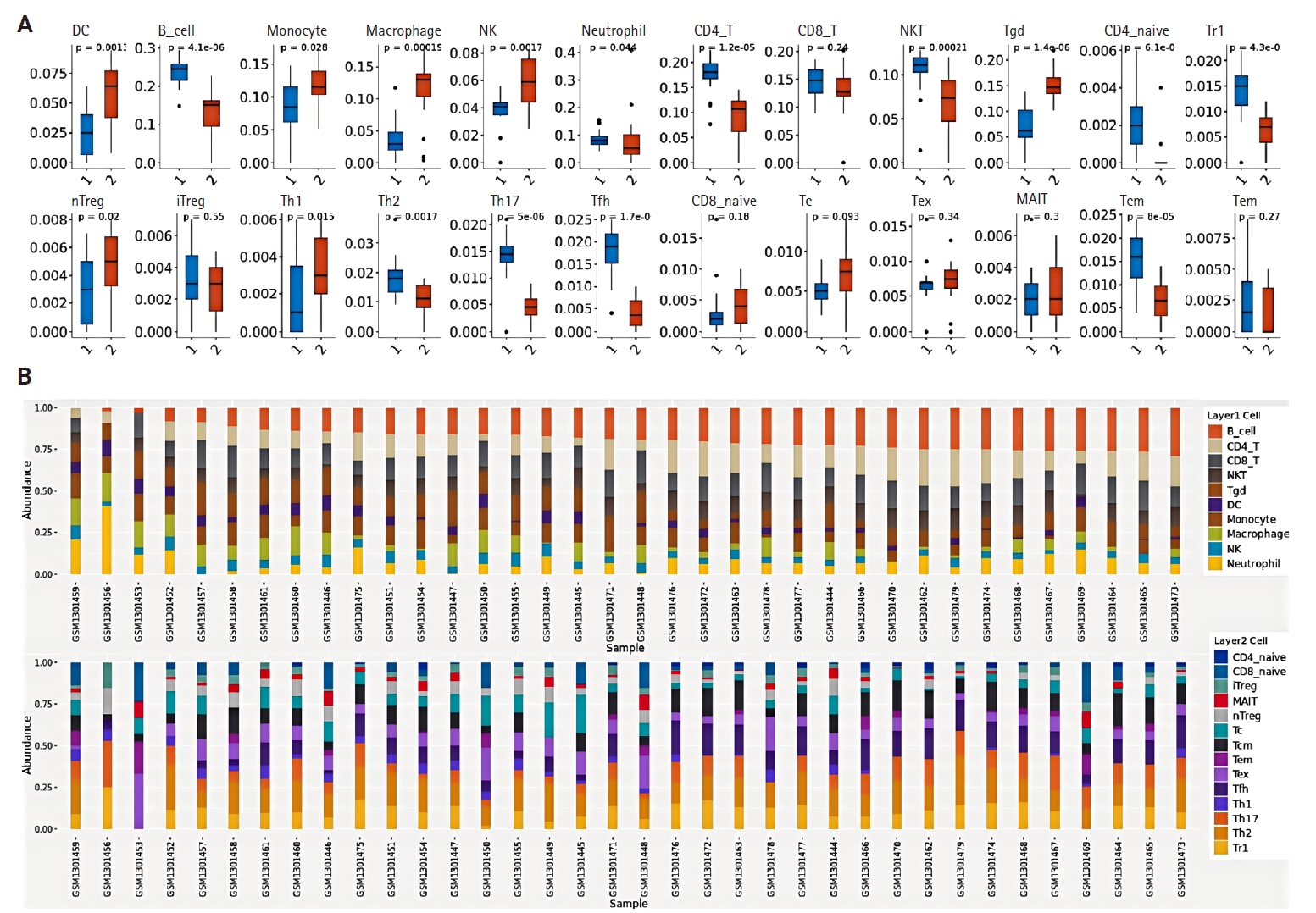
- Using the NPC dataset GSE53819, we analyzed differences in immune cell infiltration between tumor tissues and normal tissues by assessing immune cell abundance. Comparative analysis revealed significantly increased abundance of dendritic cells (DCs), monocytes, macrophages, natural killer (NK) cells, and gamma-delta T cells (γδ T cells) in NPC tissues compared to normal tissues, whereas reduced abundance was observed in regulatory type 1 T cells, T helper 17 cells, follicular helper T cells, central memory T cells, natural killer T cells (NKT), B cells, and CD4+ T cells (p < .05) (Fig. 2A). Fig. 2B illustrates the proportional distribution of distinct immune cell subtypes across tissue samples.
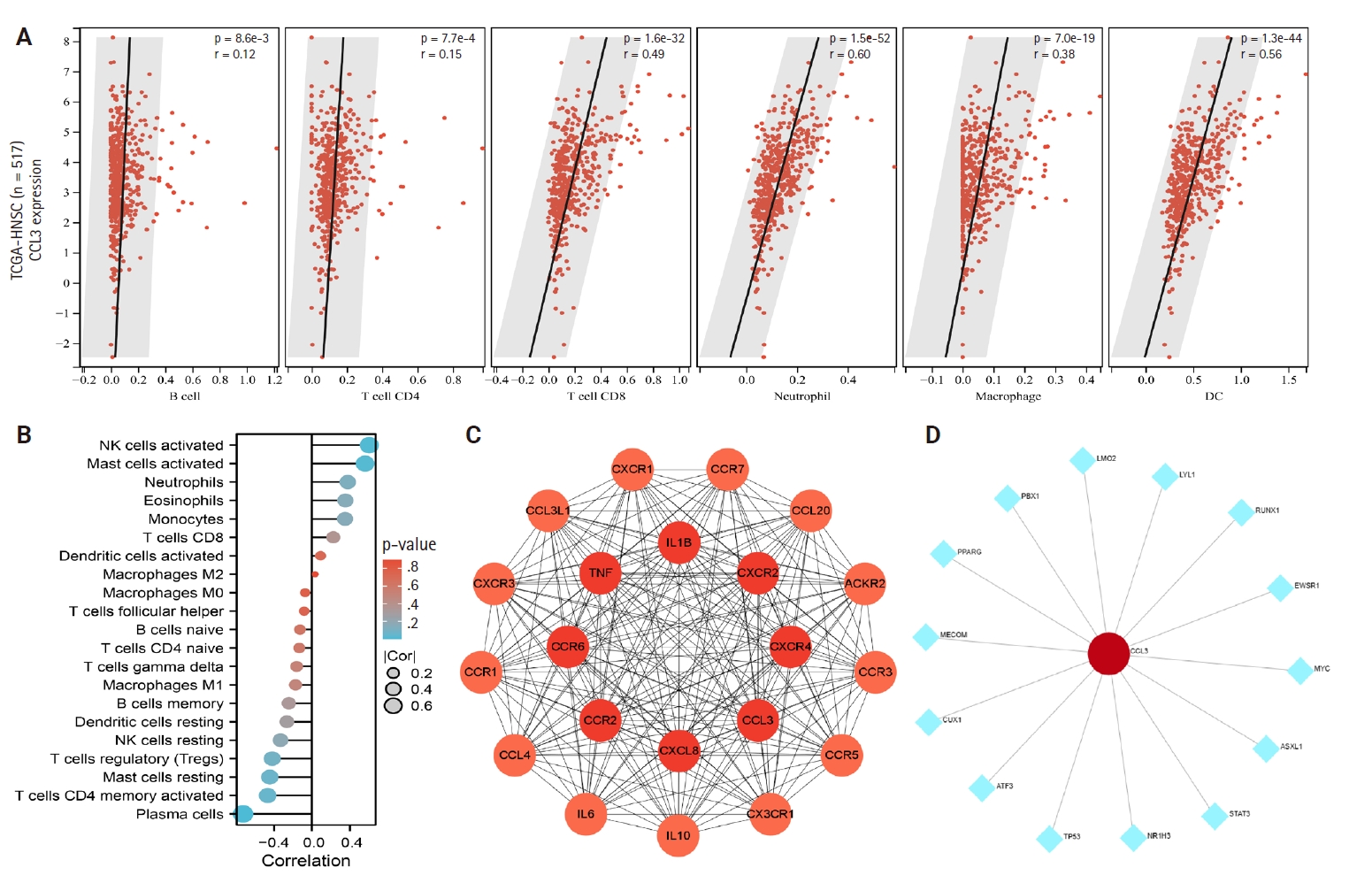
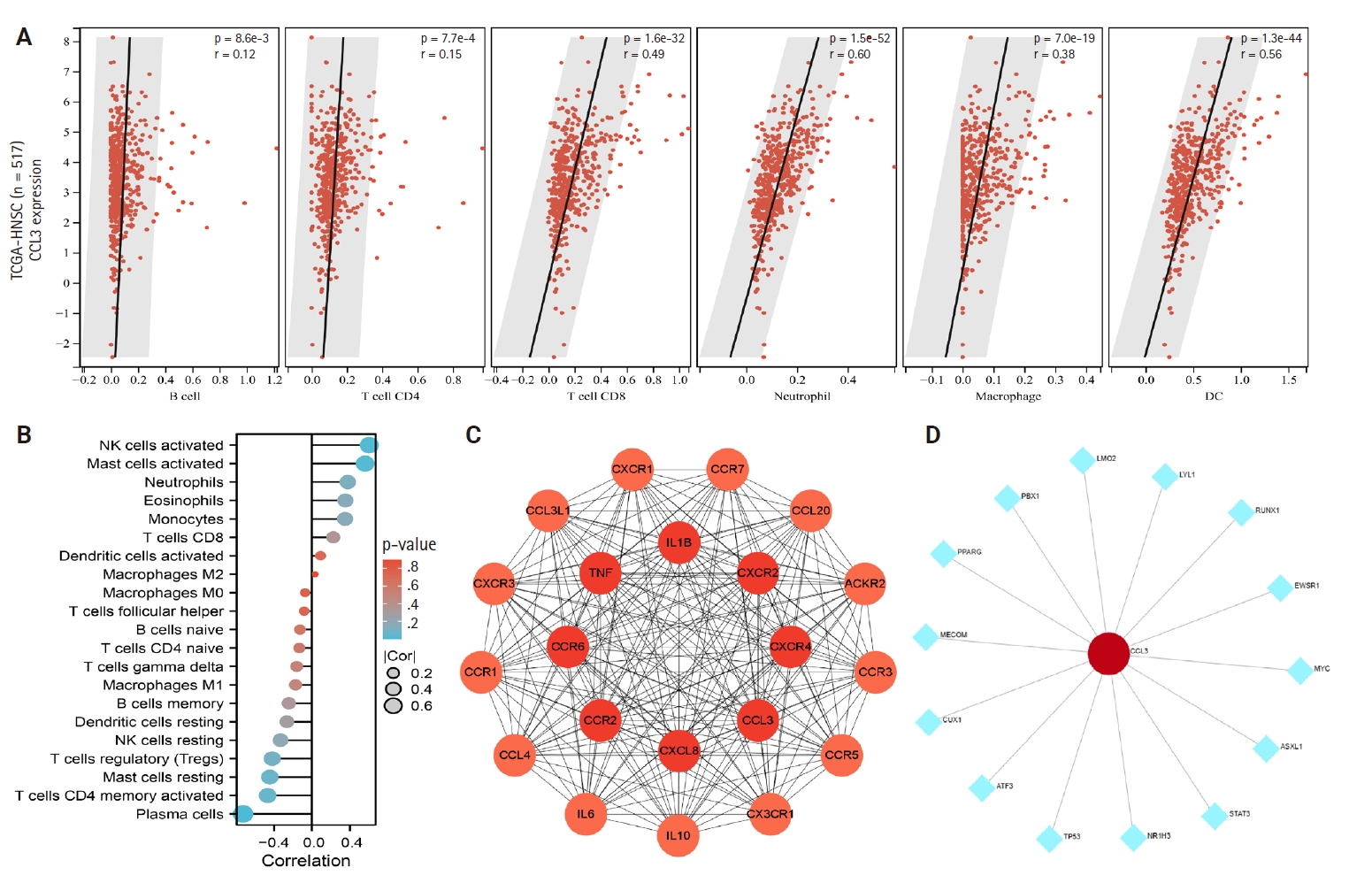
- The relationship between CCL3 expression and the infiltration of various immune cell types was analyzed using the TIMER algorithm. A positive correlation was observed between CCL3 expression and the infiltration of macrophages (r = 0.38, p < .05), CD8+ T cells (r = 0.49, p < .05), dendritic cells (r = 0.56, p < .05), and neutrophils (r = 0.60, p < .05). In contrast, B cells (r = 0.12, p < .05) and CD4+ T cells (r = 0.15, p < .05) displayed only a weak correlation with CCL3 expression (Fig. 3A).
- Based on the NPC dataset GSE53819, we employed the CIBERSORT algorithm to analyze the correlation between CCL3 expression and immune cell infiltration levels. The results revealed a significant negative correlation between CCL3 expression and plasma cells (r = –0.73, p < .001), whereas positive correlations were observed with activated NK cells (r = 0.61, p < .01) and activated mast cells (r = 0.56, p < .05) (Fig. 3B).
- Exploration of the interactions between CCL3 and other genes
- A PPI network for CCL3 and its associated genes was constructed with a confidence score of 0.40 using the STRING database. As shown in Fig. 3C, the network comprised 21 genes. To further elucidate potential target genes of CCL3, the NetworkAnalyst tool was employed, predicting 14 possible interacting genes (Fig. 3D).
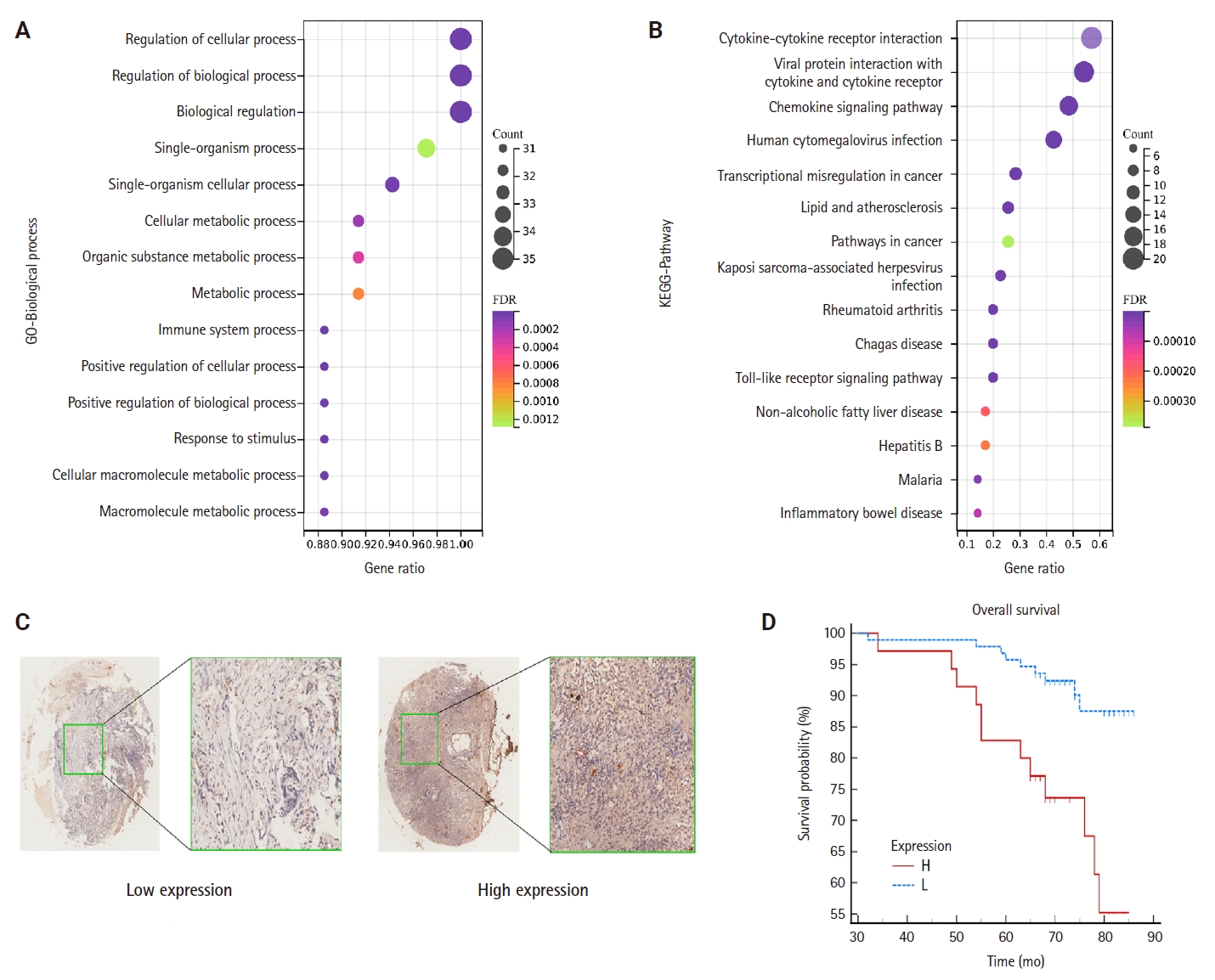
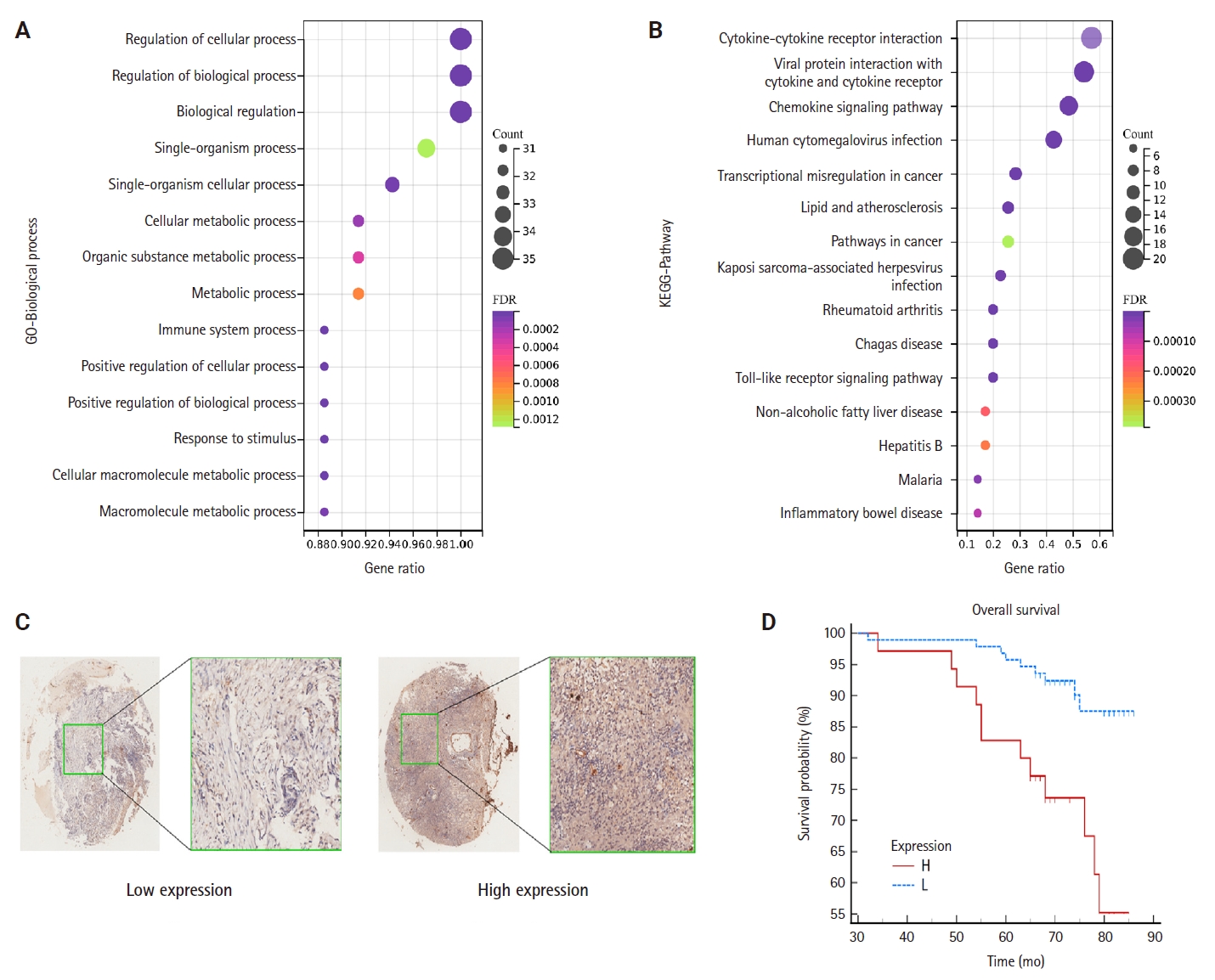
- To elucidate the potential functions of genes associated with CCL3, we performed GO and KEGG enrichment analyses. The results demonstrated that these genes were significantly enriched in 1,108 GO biological process terms and 56 pathways, highlighting their intricate roles and potential involvement in regulating cell growth through diverse mechanisms. We selected the top-ranked terms for visualization (Fig. 4A). The findings indicate that pathways such as the cancer pathway, chemokine signaling pathway, Toll-like receptor signaling pathway, immune system processes, signal transduction, and biological regulation (Fig. 4B), which were prominently ranked, may play crucial roles in the development of NPC.
- The results of the IHC staining of CCL3 based on a TMA of NPC
- TMA were employed to detect CCL3 protein expression via IHC. The analysis revealed that CCL3 was predominantly localized in the cytoplasm of tumor cells (Fig. 4C). Furthermore, Fig. 4D demonstrates that patients with elevated CCL3 expression experienced a poorer prognosis compared to those with lower CCL3 levels. As shown in Table 1, no significant correlation was observed with age and gender. Moreover, CCL3 expression was markedly elevated in samples with lymph node metastasis (LNM) and advanced clinical stages. Similarly, significantly higher levels of CCL3 were observed in samples exhibiting recurrence (Table 1).
RESULTS
PPI network
GO and KEGG enrichment analysis
- CCL3 is a pivotal chemokine, prominently expressed across various tumors, and closely linked to the tumor immune microenvironment. Elevated CCL3 expression is associated with poor prognosis in diffuse large B-cell lymphoma and colon adenocarcinoma, where it facilitates cell migration and invasion [25,26]. Nonetheless, research on CCL3 in NPC remains limited. The results of the present study demonstrated high expression of CCL3 in NPC and its correlation with poor prognosis, the presence of LNM, and cancer recurrence. These findings suggest that CCL3 may serve as a central gene in NPC, influencing crucial biological processes and pathways that drive the disease’s onset and progression.
- Notably, the expression of CCL3 was predicted to correlate with the sensitivity of cancer cells to a number of drugs. Among these, 5-fluorouracil, a key agent in the clinical treatment of NPC and a cornerstone of NPC chemotherapy, significantly enhances survival in patients with chemotherapy-sensitive NPC when combined with radiotherapy [27]. However, prolonged administration of 5-fluorouracil can lead to acquired resistance in tumor cells, thereby limiting its clinical efficacy [28]. This study found a negative correlation between high CCL3 expression and 5-fluorouracil sensitivity, indicating that elevated CCL3 levels may diminish the effectiveness of certain chemotherapeutic agents. Consequently, targeting CCL3 could potentially mitigate chemoresistance in NPC, thereby improving patient prognosis.
- The findings demonstrated distinct alterations in immune cell infiltration abundance during the transition from normal tissues to NPC tissues. Specifically, increased infiltration of DCs and NK cells was observed, whereas decreased abundance of NKT cells and CD4+ T cells was detected. These results suggest that dynamic changes in immune cell infiltration may influence NPC progression and therapeutic response. Furthermore, this study revealed that CCL3 expression was positively correlated with the infiltration of NK cells, activated mast cells, CD8+ T cells, dendritic cells, and neutrophils. Conversely, CCL3 expression showed a negative correlation with plasma cell infiltration. Notably, certain immune cells, such as NKT cells, are crucial in anti-tumor activity. NKT cells can significantly impact immune surveillance and anti-tumor immunity through both direct and indirect mechanisms [29]. The activation of NKT cells can trigger a robust anti-tumor immune response. Additionally, dendritic cells play a pivotal role in presenting antigens, which promotes tumor-specific CD8+ T-cell responses and enhances the effectiveness of CD8+ T cells in tumor immunity and tumor cell eradication [30]. However, certain studies have indicated that immune cell infiltration can also facilitate tumor progression. For instance, the cross-presentation of tumor antigens by dendritic cells in the tumor microenvironment usually induces T-cell tolerance rather than an immune response [31]. Additionally, the infiltration of neutrophils and the formation of neutrophil extracellular traps have been shown to promote tumor cell metastasis [32]. Furthermore, tumor-associated macrophages, which infiltrate tumors, can be categorized into M1 and M2 subtypes. M1 macrophages generally exhibit anti-tumor activity, while M2 macrophages are known to facilitate tumor progression and metastasis. Nonetheless, these macrophages can undergo reciprocal transformation in response to alterations in the tumor microenvironment or therapeutic interventions [33]. Additionally, research has identified two subgroups of NKT cells: type I NKT cells, which enhance anti-tumor responses, and type II NKT cells, which suppress these responses [34]. In summary, it is evident that CCL3 is intricately linked to immune cell infiltration. However, the influence of CCL3 on tumor development is attributed to a complex interplay of various mechanisms, and elucidating the specific mechanisms by which CCL3 affects immune infiltration will be the focal point of our forthcoming research.
- Through bioinformatics analysis, we have discovered that CCL3 interacts with 20 genes, including CCL2, CCR1 (C-C motif chemokine receptor 1), and TNF (tumor necrosis factor). Additionally, CCL3 may influence target genes such as PBX1, LYL1, and TP53D. These genes are predominantly involved in immune-related biological processes, including cytokine-mediated signaling pathways, monocyte chemotaxis, and immune system regulation. KEGG pathway enrichment analysis indicates that these genes are associated with several critical pathways, such as the mitogen-activated protein kinase (MAPK) signaling pathway, interleukin 17 (IL-17) signaling pathways, TNF signaling pathways, and T-cell receptor signaling pathways. These pathways are likely to play a pivotal role in the onset and progression of tumors.
- For example, research has demonstrated that Mex3a facilitates the onset of colorectal cancer via the MAPK signaling pathway [35]. Targeting the MAPK pathway can influence the chemotherapy sensitivity of cancer cells [36]. IL-17 plays a critical role in advancing cancer progression by sustaining a chronic inflammatory microenvironment conducive to tumor development [37]. These findings imply that the mechanism through which CCL3 fosters NPC development is profoundly intricate, likely involving a multitude of biological processes and signaling pathways.
- The present study may have some limitations. Firstly, the bioinformatics study of NPC is limited because there is relatively little data on this cancer in public databases. Secondly, our investigation into the potential mechanisms of CCL3 is predominantly based on bioinformatics analyses. Therefore, additional experimental studies are required to validate its role in the development of NPC in future research. Finally, the relatively limited sample size of the NPC TMA and insufficient validation assays of immune cells by IHC may introduce potential bias. Future studies will incorporate expanded cohorts and additional experimental validations to address these limitations.
- In conclusion, this study has found that CCL3 may be a key gene in the development of NPC and holds promise as both a diagnostic marker and a therapeutic target for this malignancy.
DISCUSSION
Ethics Statement
The TMA was commercially purchased, which was approved by the Institutional Review Board of Shanghai Outdo Biotech Co., Ltd. (SHYJS-CP-1810011).
Availability of Data and Material
Data are available on reasonable request. The data used to support the findings of this study are available from the corresponding author on request.
Code Availability
Not applicable.
Author Contributions
Conceptualization: XZ, JL. Data Curation: HZ, AC. Formal Analysis: XG, ZS, YL. Investigation: JL, AC. Project Administration: XZ, HZ. Methodology: XZ, HZ. Resources: JL, YL. Validation: JL, XG, ZS. Visualization: XG, ZS. Writing—original draft preparation: XG, ZS. Writing—review & editing: all authors. Approval of the final manuscript: all authors.
Conflicts of Interest
The authors declare that they have no potential conflicts of interest.
Funding Statement
This study was partially supported by the Guizhou Science and Technology Project (2021-045 and 2023-327), and the Cultivation project of Affiliated Hospital of Guizhou Medical University (I-2020-10 and gyfybsky-2021-60).
- 1. Lee HM, Okuda KS, Gonzalez FE, Patel V. Current perspectives on nasopharyngeal carcinoma. Adv Exp Med Biol 2019; 1164: 11-34. ArticlePubMed
- 2. Chen YP, Chan AT, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet 2019; 394: 64-80. ArticlePubMed
- 3. Zhang SQ, Pan SM, Liang SX, Han YS, Chen HB, Li JC. Research status and prospects of biomarkers for nasopharyngeal carcinoma in the era of high‑throughput omics (review). Int J Oncol 2021; 58: 9.ArticlePMC
- 4. Mnejja W, Nouri O, Fourati N, et al. Current management and perspectives for locally advanced nasopharyngeal carcinoma. Cancer Radiother 2022; 26: 730-5. ArticlePubMed
- 5. Langendijk JA, Doornaert P, Verdonck-de Leeuw IM, Leemans CR, Aaronson NK, Slotman BJ. Impact of late treatment-related toxicity on quality of life among patients with head and neck cancer treated with radiotherapy. J Clin Oncol 2008; 26: 3770-6. ArticlePubMed
- 6. Zhang P, He Q, Wang Y, et al. Protein C receptor maintains cancer stem cell properties via activating lipid synthesis in nasopharyngeal carcinoma. Signal Transduct Target Ther 2022; 7: 46.ArticlePubMedPMCPDF
- 7. Ling J, Chang A, Zhao H, Ye H, Zhuo X. EPHB2 as a recurrence-related gene and a prognostic indicator in nasopharyngeal carcinoma: a bioinformatics screening and immunohistochemistry verification. Histol Histopathol 2022; 37: 889-97. ArticlePubMed
- 8. Li JY, Zhao Y, Gong S, et al. TRIM21 inhibits irradiation-induced mitochondrial DNA release and impairs antitumour immunity in nasopharyngeal carcinoma tumour models. Nat Commun 2023; 14: 865.ArticlePubMedPMCPDF
- 9. Makowska A, Lelabi N, Nothbaum C, et al. Radiotherapy combined with PD-1 inhibition increases NK cell cytotoxicity towards nasopharyngeal carcinoma cells. Cells 2021; 10: 2458.ArticlePubMedPMC
- 10. Yuan JJ, Ding JW, Li JW, et al. Nimotuzumab plus induction chemotherapy followed by radiotherapy/concurrent chemoradiotherapy plus nimotuzumab for locally advanced nasopharyngeal carcinoma: protocol of a multicentre, open-label, single-arm, prospective phase II trial. BMJ Open 2022; 12: e051594. ArticlePubMedPMC
- 11. Ntanasis-Stathopoulos I, Fotiou D, Terpos E. CCL3 signaling in the tumor microenvironment. Adv Exp Med Biol 2020; 1231: 13-21. ArticlePubMed
- 12. Ma X, Su J, Zhao S, et al. CCL3 promotes proliferation of colorectal cancer related with TRAF6/NF-kappaB molecular pathway. Contrast Media Mol Imaging 2022; 2022: 2387192.ArticlePubMedPMC
- 13. Ding L, Li B, Zhao Y, et al. Serum CCL2 and CCL3 as potential biomarkers for the diagnosis of oral squamous cell carcinoma. Tumour Biol 2014; 35: 10539-46. ArticlePubMedPDF
- 14. Bao YN, Cao X, Luo DH, et al. Urokinase-type plasminogen activator receptor signaling is critical in nasopharyngeal carcinoma cell growth and metastasis. Cell Cycle 2014; 13: 1958-69. ArticlePubMedPMC
- 15. Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013; 41: D955-61. ArticlePubMed
- 16. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017; 77: e108-10. ArticlePubMedPMCPDF
- 17. Miao YR, Zhang Q, Lei Q, et al. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinh) 2020; 7: 1902880.ArticlePubMedPMCPDF
- 18. Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021; 49: D605-12. ArticlePubMedPDF
- 19. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498-504. ArticlePubMedPMC
- 20. Zhou G, Soufan O, Ewald J, Hancock RE, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 2019; 47: W234-41. ArticlePubMedPMC
- 21. Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003; 4: P3.ArticlePubMedPDF
- 22. Jawhar NM. Tissue microarray: a rapidly evolving diagnostic and research tool. Ann Saudi Med 2009; 29: 123-7. ArticlePubMedPMC
- 23. Ling J, Zhang L, Chang A, et al. Overexpression of KITLG predicts unfavorable clinical outcomes and promotes lymph node metastasis via the JAK/STAT pathway in nasopharyngeal carcinoma. Lab Invest 2022; 102: 1257-67. ArticlePubMedPDF
- 24. Schoonjans F, Zalata A, Depuydt CE, Comhaire FH. MedCalc: a new computer program for medical statistics. Comput Methods Programs Biomed 1995; 48: 257-62. ArticlePubMed
- 25. Takahashi K, Sivina M, Hoellenriegel J, et al. CCL3 and CCL4 are biomarkers for B cell receptor pathway activation and prognostic serum markers in diffuse large B cell lymphoma. Br J Haematol 2015; 171: 726-35. ArticlePubMedPMCPDF
- 26. Guan B, Li H, Yao J, et al. CCL3-CCR5 axis promotes cell migration and invasion of colon adenocarcinoma via Akt signaling pathway. Environ Toxicol 2023; 38: 172-84. ArticlePubMed
- 27. You R, Liu YP, Huang PY, et al. Efficacy and safety of locoregional radiotherapy with chemotherapy vs chemotherapy alone in de novo metastatic nasopharyngeal carcinoma: a multicenter phase 3 randomized clinical trial. JAMA Oncol 2020; 6: 1345-52. ArticlePubMedPMC
- 28. Feng X, Luo Q, Zhang H, et al. The role of NLRP3 inflammasome in 5-fluorouracil resistance of oral squamous cell carcinoma. J Exp Clin Cancer Res 2017; 36: 81.ArticlePubMedPMCPDF
- 29. Nelson A, Lukacs JD, Johnston B. The current landscape of NKT cell immunotherapy and the hills ahead. Cancers (Basel) 2021; 13: 5174.ArticlePubMedPMC
- 30. MacNabb BW, Tumuluru S, Chen X, et al. Dendritic cells can prime anti-tumor CD8(+) T cell responses through major histocompatibility complex cross-dressing. Immunity 2022; 55: 982-97. ArticlePubMedPMC
- 31. Fu C, Jiang A. Dendritic cells and CD8 T cell immunity in tumor microenvironment. Front Immunol 2018; 9: 3059.ArticlePubMedPMC
- 32. Xiao Y, Cong M, Li J, et al. Cathepsin C promotes breast cancer lung metastasis by modulating neutrophil infiltration and neutrophil extracellular trap formation. Cancer Cell 2021; 39: 423-37. ArticlePubMed
- 33. Pan Y, Yu Y, Wang X, Zhang T. Tumor-associated macrophages in tumor immunity. Front Immunol 2020; 11: 583084.ArticlePubMedPMC
- 34. Terabe M, Berzofsky JA. NKT cells in immunoregulation of tumor immunity: a new immunoregulatory axis. Trends Immunol 2007; 28: 491-6. ArticlePubMed
- 35. Li H, Liang J, Wang J, et al. Mex3a promotes oncogenesis through the RAP1/MAPK signaling pathway in colorectal cancer and is inhibited by hsa-miR-6887-3p. Cancer Commun (Lond) 2021; 41: 472-91. ArticlePubMedPMCPDF
- 36. Lee S, Rauch J, Kolch W. Targeting MAPK signaling in cancer: mechanisms of drug resistance and sensitivity. Int J Mol Sci 2020; 21: 1102.ArticlePubMedPMC
- 37. Wu L, Chen X, Zhao J, et al. A novel IL-17 signaling pathway controlling keratinocyte proliferation and tumorigenesis via the TRAF4-ERK5 axis. J Exp Med 2015; 212: 1571-87. ArticlePubMedPMCPDF
REFERENCES
Figure & Data
References
Citations

 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure
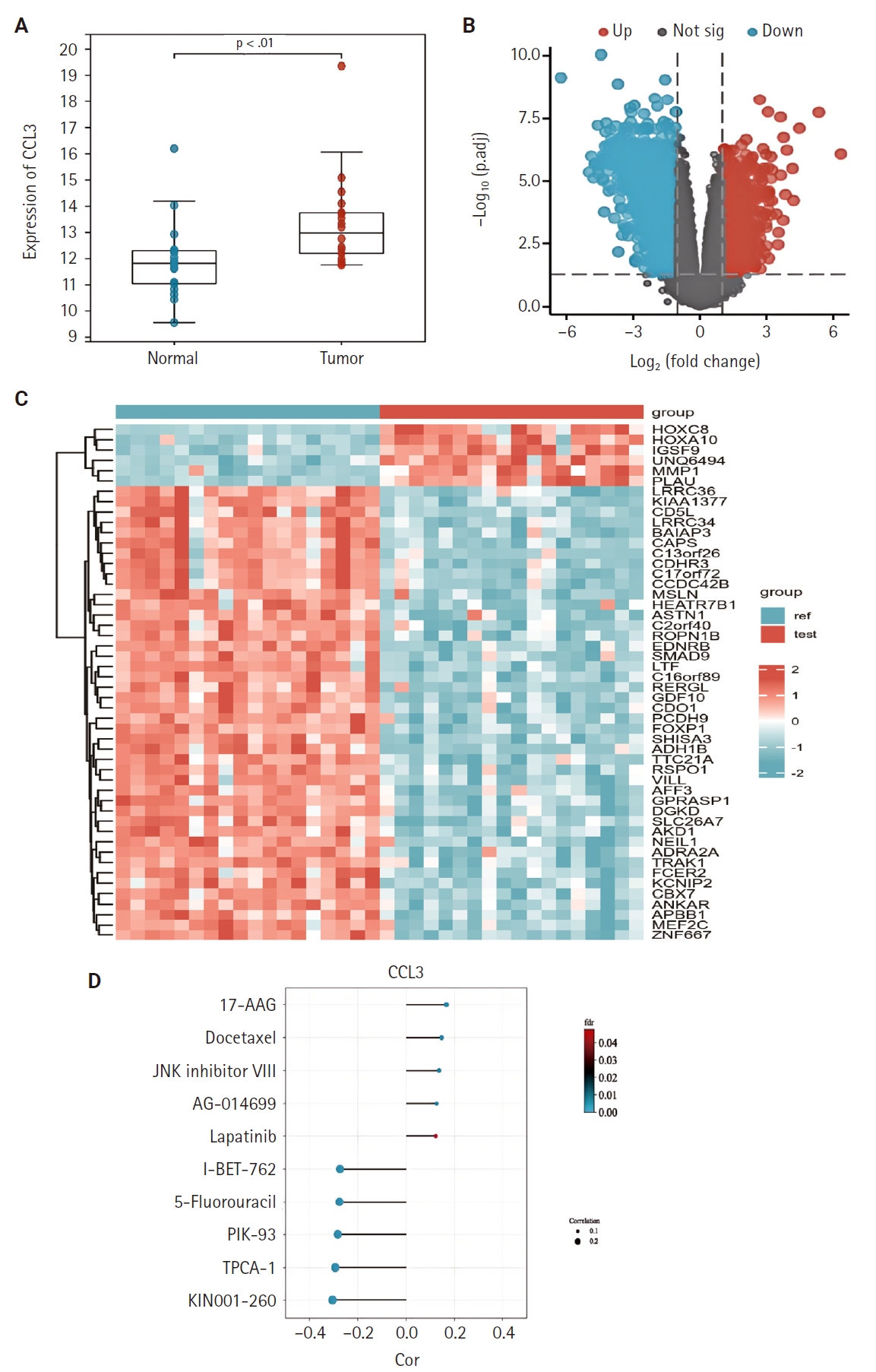
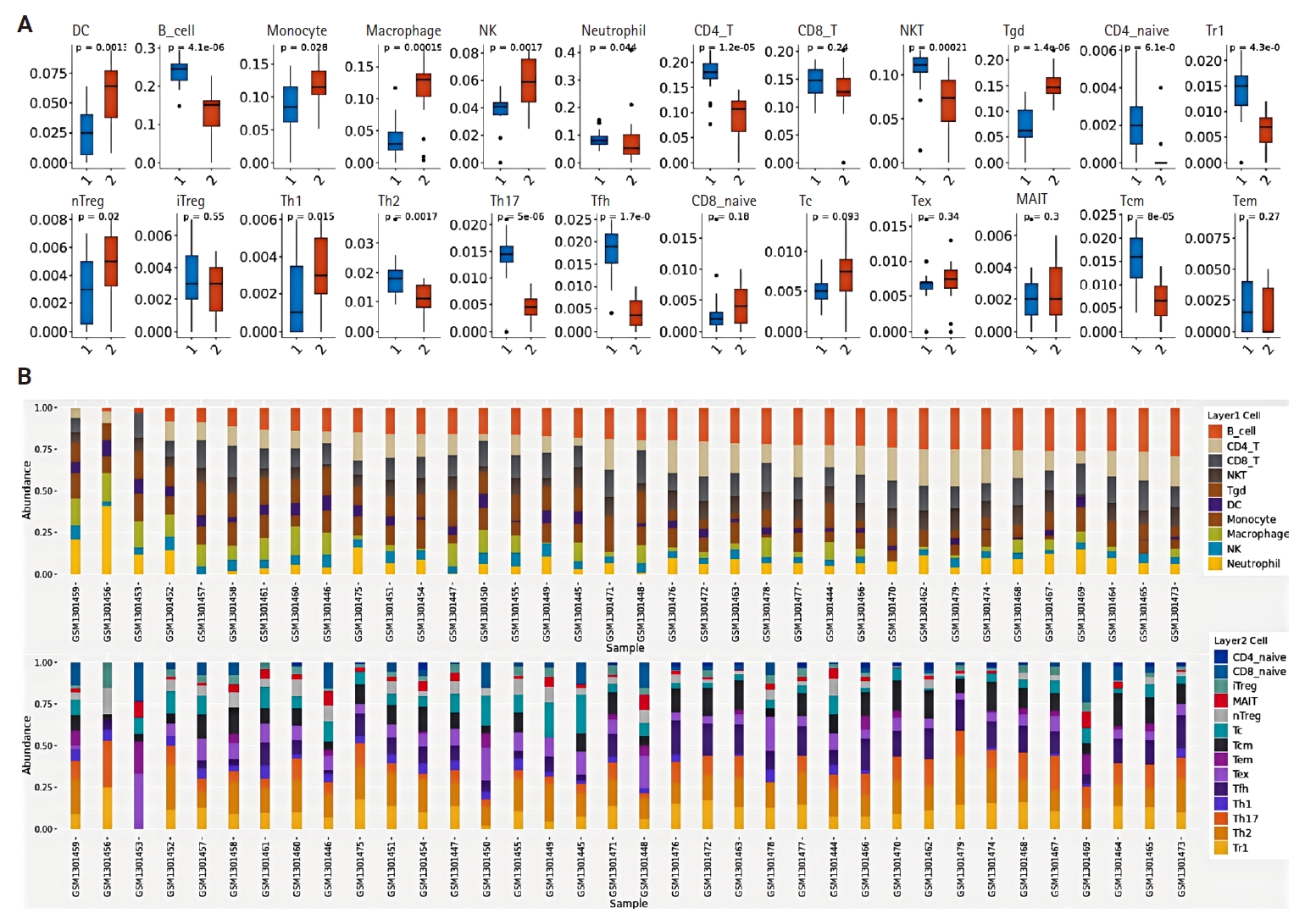
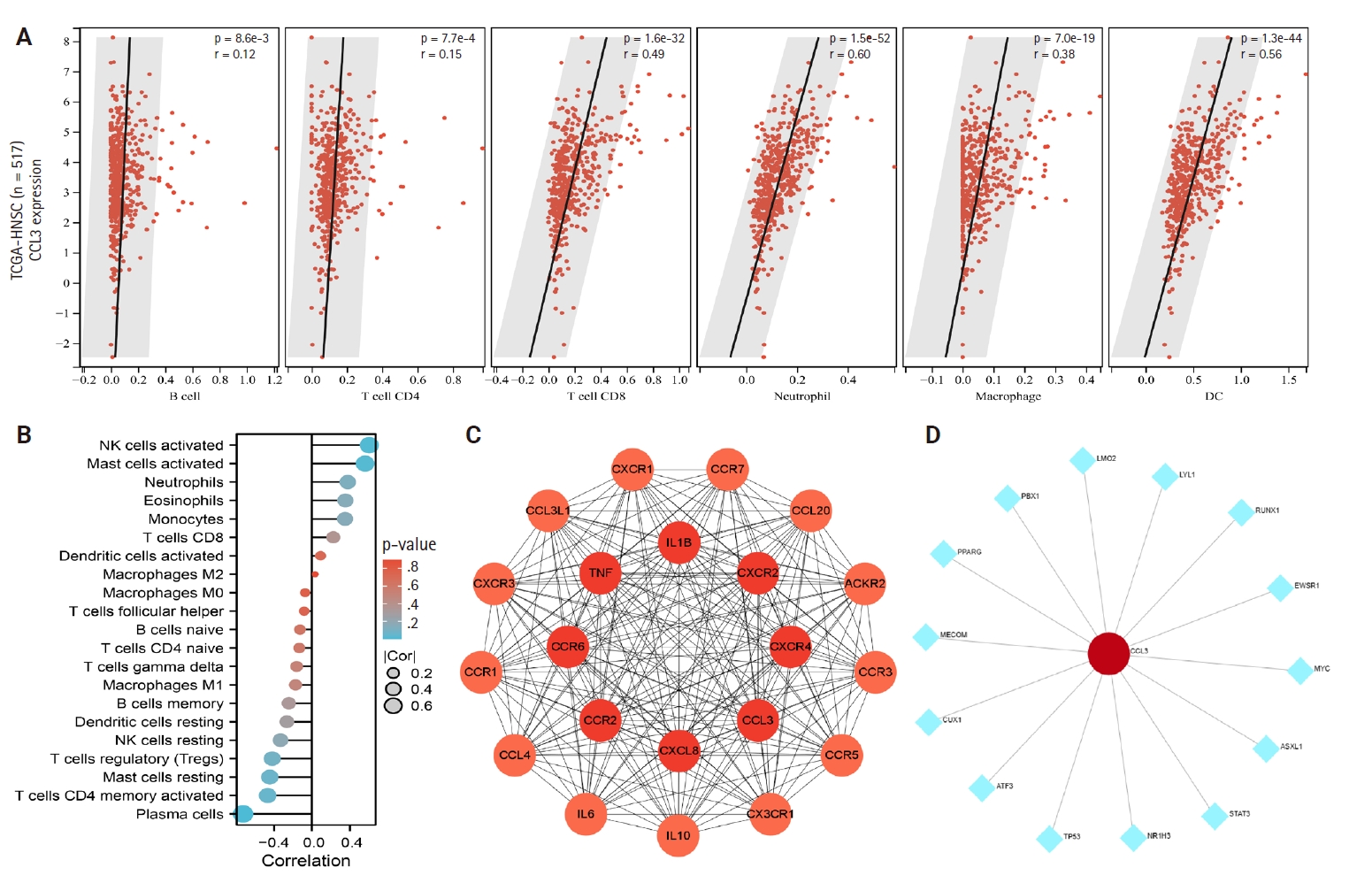
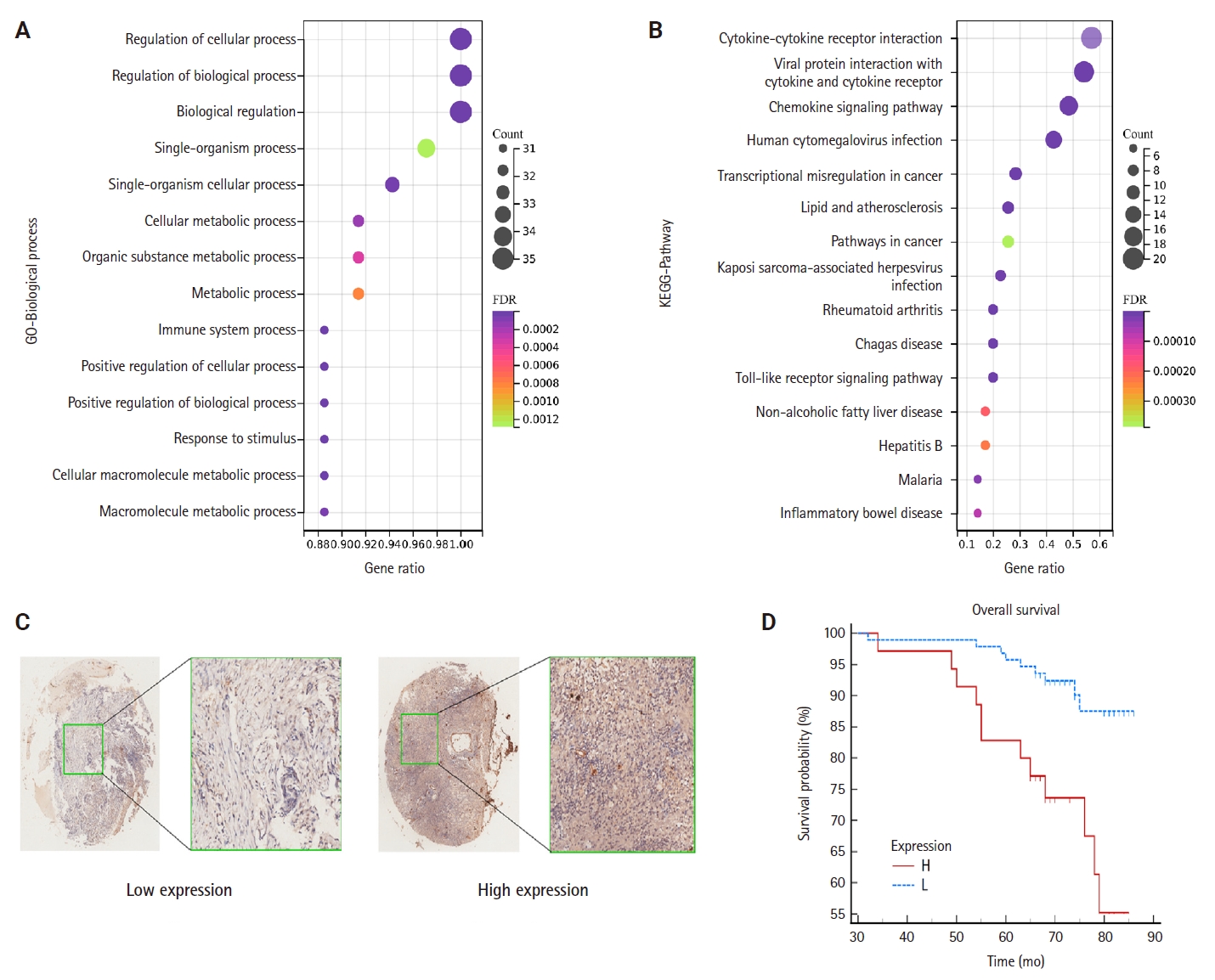
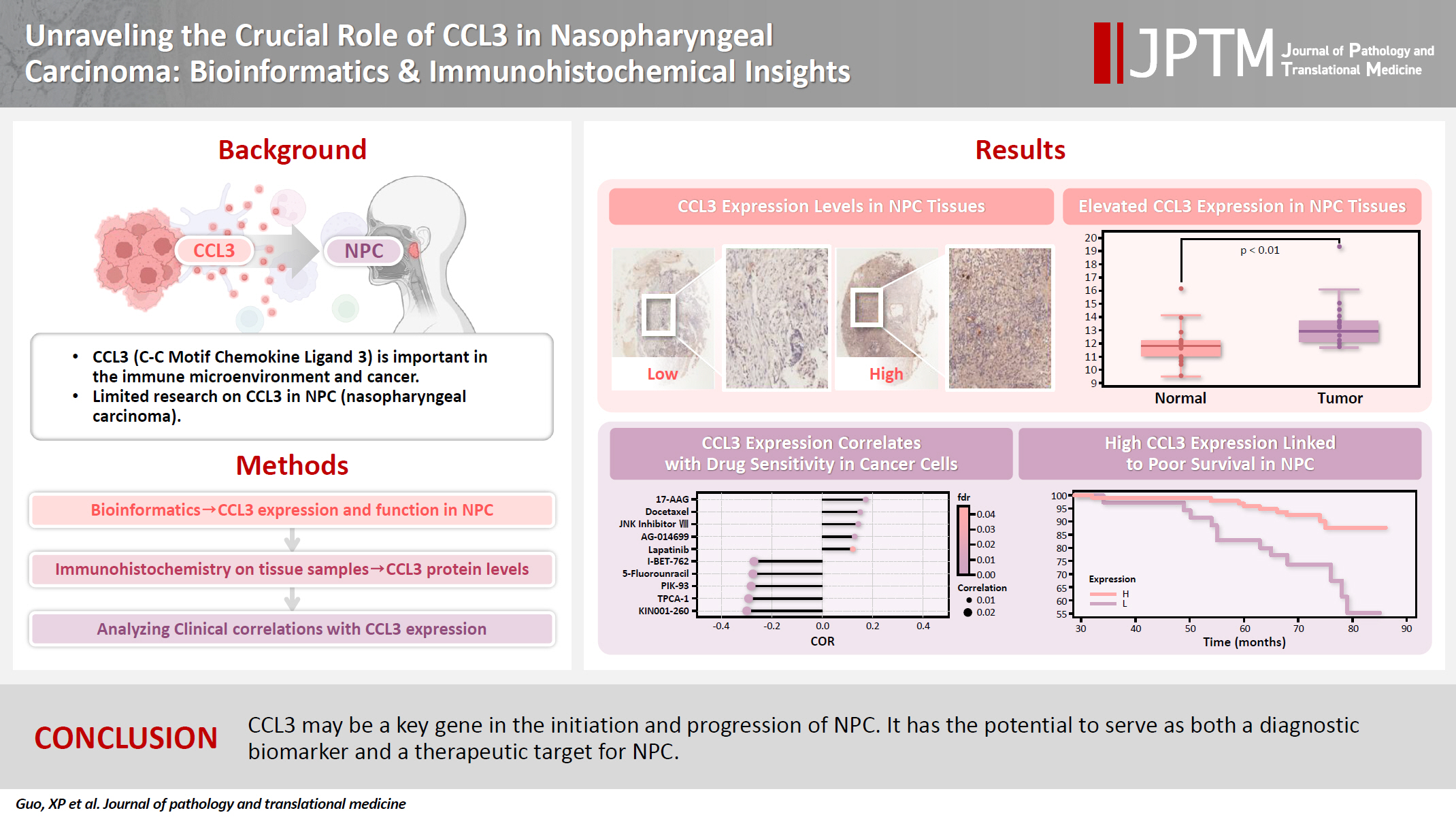
Fig. 1.
Fig. 2.
Fig. 3.
Fig. 4.
Graphical abstract
| Variable | Total | CCL3 expression |
p-value | |
|---|---|---|---|---|
| High | Low | |||
| Age (yr) | ||||
| <50 | 72 | 20 (27.8) | 52 (72.2) | .380 |
| ≥50 | 57 | 12 (21.0) | 45 (79.0) | |
| Sex | ||||
| Male | 99 | 22 (22.2) | 77 (77.8) | .217 |
| Female | 30 | 10 (33.3) | 20 (66.7) | |
| Recurrence | ||||
| Yes | 59 | 22 (37.3) | 37 (62.7) | .001 |
| No | 69 | 9 (13.0) | 60 (87.0) | |
| Clinical stage | ||||
| 1 + 2 | 70 | 11 (15.7) | 59 (84.3) | .009 |
| 3 + 4 | 59 | 21 (35.6) | 38 (64.4) | |
| Neck LNM | ||||
| With | 93 | 28 (30.1) | 65 (69.9) | .025 |
| Without | 36 | 4 (11.1) | 32 (88.9) | |
CCL3, C-C motif chemokine ligand 3; LNM, lymph node metastasis.

 E-submission
E-submission