Search
- Page Path
- HOME > Search
- KRAS Mutation Test in Korean Patients with Colorectal Carcinomas: A Methodological Comparison between Sanger Sequencing and a Real-Time PCR-Based Assay
- Sung Hak Lee, Arthur Minwoo Chung, Ahwon Lee, Woo Jin Oh, Yeong Jin Choi, Youn-Soo Lee, Eun Sun Jung
- J Pathol Transl Med. 2017;51(1):24-31. Published online December 25, 2016
- DOI: https://doi.org/10.4132/jptm.2016.10.03
- 12,404 View
- 170 Download
- 5 Web of Science
- 5 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
Mutations in the KRAS gene have been identified in approximately 50% of colorectal cancers (CRCs). KRAS mutations are well established biomarkers in anti–epidermal growth factor receptor therapy. Therefore, assessment of KRAS mutations is needed in CRC patients to ensure appropriate treatment.
Methods
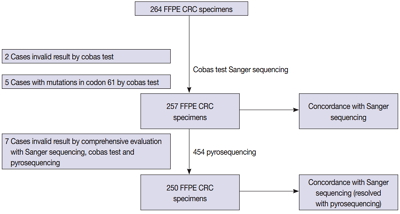
We compared the analytical performance of the cobas test to Sanger sequencing in 264 CRC cases. In addition, discordant specimens were evaluated by 454 pyrosequencing.
Results
KRAS mutations for codons 12/13 were detected in 43.2% of cases (114/264) by Sanger sequencing. Of 257 evaluable specimens for comparison, KRAS mutations were detected in 112 cases (43.6%) by Sanger sequencing and 118 cases (45.9%) by the cobas test. Concordance between the cobas test and Sanger sequencing for each lot was 93.8% positive percent agreement (PPA) and 91.0% negative percent agreement (NPA) for codons 12/13. Results from the cobas test and Sanger sequencing were discordant for 20 cases (7.8%). Twenty discrepant cases were subsequently subjected to 454 pyrosequencing. After comprehensive analysis of the results from combined Sanger sequencing–454 pyrosequencing and the cobas test, PPA was 97.5% and NPA was 100%.
Conclusions
The cobas test is an accurate and sensitive test for detecting KRAS-activating mutations and has analytical power equivalent to Sanger sequencing. Prescreening using the cobas test with subsequent application of Sanger sequencing is the best strategy for routine detection of KRAS mutations in CRC. -
Citations
Citations to this article as recorded by- Single-center study on clinicopathological and typical molecular pathologic features of metastatic brain tumor
Su Hwa Kim, Young Suk Lee, Sung Hak Lee, Yeoun Eun Sung, Ahwon Lee, Jun Kang, Jae-Sung Park, Sin Soo Jeun, Youn Soo Lee
Journal of Pathology and Translational Medicine.2023; 57(4): 217. CrossRef - Assessment of KRAS and NRAS status in metastatic colorectal cancer: Experience of the National Institute of Oncology in Rabat Morocco
Chaimaa Mounjid, Hajar El Agouri, Youssef Mahdi, Abdelilah Laraqui, En-nacer Chtati, Soumaya Ech-charif, Mouna Khmou, Youssef Bakri, Amine Souadka, Basma El Khannoussi
Annals of Cancer Research and Therapy.2022; 30(2): 80. CrossRef - The current understanding on the impact of KRAS on colorectal cancer
Mingjing Meng, Keying Zhong, Ting Jiang, Zhongqiu Liu, Hiu Yee Kwan, Tao Su
Biomedicine & Pharmacotherapy.2021; 140: 111717. CrossRef - Droplet digital PCR revealed high concordance between primary tumors and lymph node metastases in multiplex screening of KRAS mutations in colorectal cancer
Barbora Vanova, Michal Kalman, Karin Jasek, Ivana Kasubova, Tatiana Burjanivova, Anna Farkasova, Peter Kruzliak, Dietrich Busselberg, Lukas Plank, Zora Lasabova
Clinical and Experimental Medicine.2019; 19(2): 219. CrossRef - CRISPR Technology for Breast Cancer: Diagnostics, Modeling, and Therapy
Rachel L. Mintz, Madeleine A. Gao, Kahmun Lo, Yeh‐Hsing Lao, Mingqiang Li, Kam W. Leong
Advanced Biosystems.2018;[Epub] CrossRef
- Single-center study on clinicopathological and typical molecular pathologic features of metastatic brain tumor
- Comparison of Three
BRAF Mutation Tests in Formalin-Fixed Paraffin Embedded Clinical Samples - Soomin Ahn, Jeeyun Lee, Ji-Youn Sung, So Young Kang, Sang Yun Ha, Kee-Taek Jang, Yoon-La Choi, Jung-Sun Kim, Young Lyun Oh, Kyoung-Mee Kim
- Korean J Pathol. 2013;47(4):348-354. Published online August 26, 2013
- DOI: https://doi.org/10.4132/KoreanJPathol.2013.47.4.348
- 9,874 View
- 60 Download
- 9 Crossref
-
 Abstract
Abstract
 PDF
PDF Background Recently,
BRAF inhibitors showed dramatic treatment outcomes inBRAF V600 mutant melanoma. Therefore, the accuracy ofBRAF mutation test is critical.Methods BRAF mutations were tested by dual-priming oligonucleotide-polymerase chain reaction (DPO-PCR), direct sequencing and subsequently retested with a real-time PCR assay, cobas 4800 V600 mutation test. In total, 64 tumors including 34 malignant melanomas and 16 papillary thyroid carcinomas were analyzed. DNA was extracted from formalin-fixed paraffin embedded tissue samples and the results of cobas test were directly compared with those of DPO-PCR and direct sequencing.Results BRAF mutations were found in 23 of 64 (35.9%) tumors. There was 9.4% discordance among 3 methods. Out of 6 discordant cases, 4 cases were melanomas; 3 cases wereBRAF V600E detected only by cobas test, but were not detected by DPO-PCR and direct sequencing. One melanoma patient withBRAF mutation detected only by cobas test has been on vemurafenib treatment for 6 months and showed a dramatic response to vemurafenib. DPO-PCR failed to detect V600K mutation in one case identified by both direct sequencing and cobas test.Conclusions In direct comparison of the currently available DPO-PCR, direct sequencing and real-time cobas test for
BRAF mutation, real-time PCR assay is the most sensitive method.-
Citations
Citations to this article as recorded by- Preoperative BRAFV600E mutation detection in thyroid carcinoma by immunocytochemistry
Kristine Zøylner Swan, Stine Horskær Madsen, Steen Joop Bonnema, Viveque Egsgaard Nielsen, Marie Louise Jespersen
APMIS.2022; 130(11): 627. CrossRef - Strategy to reduce unnecessary surgeries in thyroid nodules with cytology of Bethesda category III (AUS/FLUS): a retrospective analysis of 667 patients diagnosed by surgery
Yong Joon Suh, Yeon Ju Choi
Endocrine.2020; 69(3): 578. CrossRef - A new primer construction technique that effectively increases amplification of rare mutant templates in samples
Jr-Kai Huang, Ling Fan, Tao-Yeuan Wang, Pao-Shu Wu
BMC Biotechnology.2019;[Epub] CrossRef - BRAF and NRAS mutations and antitumor immunity in Korean malignant melanomas and their prognostic relevance: Gene set enrichment analysis and CIBERSORT analysis
Kyueng-Whan Min, Ji-Young Choe, Mi Jung Kwon, Hye Kyung Lee, Ho Suk Kang, Eun Sook Nam, Seong Jin Cho, Hye-Rim Park, Soo Kee Min, Jinwon Seo, Yun Joong Kim, Nan Young Kim, Ho Young Kim
Pathology - Research and Practice.2019; 215(12): 152671. CrossRef - The association between dermoscopic features and BRAF mutational status in cutaneous melanoma: Significance of the blue-white veil
Miquel Armengot-Carbó, Eduardo Nagore, Zaida García-Casado, Rafael Botella-Estrada
Journal of the American Academy of Dermatology.2018; 78(5): 920. CrossRef - Comparison of Five Different Assays for the Detection of BRAF Mutations in Formalin-Fixed Paraffin Embedded Tissues of Patients with Metastatic Melanoma
Claire Franczak, Julia Salleron, Cindy Dubois, Pierre Filhine-Trésarrieu, Agnès Leroux, Jean-Louis Merlin, Alexandre Harlé
Molecular Diagnosis & Therapy.2017; 21(2): 209. CrossRef - Validation of an NGS mutation detection panel for melanoma
Anne Reiman, Hugh Kikuchi, Daniela Scocchia, Peter Smith, Yee Wah Tsang, David Snead, Ian A Cree
BMC Cancer.2017;[Epub] CrossRef - Transformation to Small Cell Lung Cancer of Pulmonary Adenocarcinoma: Clinicopathologic Analysis of Six Cases
Soomin Ahn, Soo Hyun Hwang, Joungho Han, Yoon-La Choi, Se-Hoon Lee, Jin Seok Ahn, Keunchil Park, Myung-Ju Ahn, Woong-Yang Park
Journal of Pathology and Translational Medicine.2016; 50(4): 258. CrossRef - Immunohistochemistry with the anti-BRAF V600E (VE1) antibody: impact of pre-analytical conditions and concordance with DNA sequencing in colorectal and papillary thyroid carcinoma
Katerina Dvorak, Birte Aggeler, John Palting, Penny McKelvie, Andrew Ruszkiewicz, Paul Waring
Pathology.2014; 46(6): 509. CrossRef
- Preoperative BRAFV600E mutation detection in thyroid carcinoma by immunocytochemistry

 E-submission
E-submission

 First
First Prev
Prev



