Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 47(4); 2013 > Article
-
Original Article
Comparison of ThreeBRAF Mutation Tests in Formalin-Fixed Paraffin Embedded Clinical Samples - Soomin Ahn, Jeeyun Lee1, Ji-Youn Sung2, So Young Kang, Sang Yun Ha, Kee-Taek Jang, Yoon-La Choi, Jung-Sun Kim, Young Lyun Oh, Kyoung-Mee Kim
-
Korean Journal of Pathology 2013;47(4):348-354.
DOI: https://doi.org/10.4132/KoreanJPathol.2013.47.4.348
Published online: August 26, 2013
Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
1Division of Hematology-Oncology, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
2Department of Pathology, Kyung Hee University School of Medicine, Seoul, Korea.
-
Corresponding Author: Kyoung-Mee Kim, M.D. Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, 81 Irwon-ro, Gangnam-gu, Seoul 135-710, Korea. Tel: +82-2-3410-2800, Fax: +82-2-3410-0025, kkmkys@skku.edu
Corresponding Author: Jeeyun Lee, M.D. Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, 81 Irwon-ro, Gangnam-gu, Seoul 135-710, Korea. Tel: +82-2-3410-3459, Fax: +82-2-3410-0025, jyunlee@skku.edu
© 2013 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- Recently, BRAF inhibitors showed dramatic treatment outcomes in BRAF V600 mutant melanoma. Therefore, the accuracy of BRAF mutation test is critical.
-
Methods
- BRAF mutations were tested by dual-priming oligonucleotide-polymerase chain reaction (DPO-PCR), direct sequencing and subsequently retested with a real-time PCR assay, cobas 4800 V600 mutation test. In total, 64 tumors including 34 malignant melanomas and 16 papillary thyroid carcinomas were analyzed. DNA was extracted from formalin-fixed paraffin embedded tissue samples and the results of cobas test were directly compared with those of DPO-PCR and direct sequencing.
-
Results
- BRAF mutations were found in 23 of 64 (35.9%) tumors. There was 9.4% discordance among 3 methods. Out of 6 discordant cases, 4 cases were melanomas; 3 cases were BRAF V600E detected only by cobas test, but were not detected by DPO-PCR and direct sequencing. One melanoma patient with BRAF mutation detected only by cobas test has been on vemurafenib treatment for 6 months and showed a dramatic response to vemurafenib. DPO-PCR failed to detect V600K mutation in one case identified by both direct sequencing and cobas test.
-
Conclusions
- In direct comparison of the currently available DPO-PCR, direct sequencing and real-time cobas test for BRAF mutation, real-time PCR assay is the most sensitive method.
- Patients and tumors
- A total of 73 tumors were analyzed for the BRAF mutation. Tests were performed on formalin-fixed, paraffin-embedded (FFPE) tissue blocks diagnosed between 2011 and 2012. Of the 73 cases, 64 cases were sufficient for DNA isolation and analysis of the BRAF mutation by DPO-PCR, direct sequencing, and the cobas BRAF V600 mutation test, and were selected for this study. Tumor specimens consisted of 34 malignant melanomas (53.1%), 16 papillary thyroid carcinomas (25.0%), 11 adenocarcinomas from the colon (17.2%), 1 clear cell carcinoma from the skin (1.6%), 1 mucinous adenocarcinoma from the bladder (1.6%), and 1 epithelioid sarcoma (1.6%) from the soft tissue.
- Of the 34 malignant melanoma patients, 1 patient (2.9%) was stage II, 5 patients (14.3%) were stage III, 26 patients (76.5%) were stage IV, and 2 patients (5.9%) were of unknown stage. In terms of primary sites, 12 (35.3%) were cutaneous, 8 (23.5%) were mucosal, 12 (35.3%) were acral, and 2 (5.9%) were of unknown primary site. Of the 34 melanomas, 13 (38.2%) specimens were obtained from primary tumor sites and 21 (61.8%) were obtained from metastatic tumors. For the rest of the tumors, the tumor samples for BRAF detection were obtained either from primary or from metastatic tumors.
- DNA extraction
- An appropriate paraffin block containing tumor tissue was selected for analysis after reviewing the hematoxylin and eosin-stained slides. Tumor volume was evaluated for each case and the amounts of melanin pigments, necrosis, and inflammatory cells admixed with tumor were also evaluated for melanoma cases. If possible, tumor-rich areas (>80%) were extracted from FFPE tumor samples using manual microdissection under microscopy, as previously described.14 Five sections of 4 µm thickness containing a representative portion of each tumor were subjected to DNA isolation by QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol after deparaffinization with xylene. The same DNA was used for all of the analyses.
- Mutation analyses
- BRAF mutations were detected utilizing both DPO-PCR and direct sequencing. Afterwards, all of the DNA was subsequently retested with a real-time PCR assay (cobas 4800 V600 mutation test, Roche Molecular Systems, Mannheim, Germany). An agreement analysis among DPO-PCR, direct sequencing, and cobas was made.
- DPO-PCR analysis was performed using the Seeplex BRAF ACE detection system (Seegene, Seoul, Korea) designed to detect the V600E variant according to the manufacturer's instructions. Each PCR reaction mixture contained 4 µL of 5× BRAF primer, 3 µL of extracted DNA (10 ng/µL), 3 µL of 8-methoxypsoralen solution (0.01%) , and 10 µL of 2× multiplex master mix (Seegene), with a total volume of 20 µL. After 15 minutes of incubation at 94℃, amplification was performed in a Thermal Cycler (Applied Biosystems, Foster City, CA, USA) with 35 cycles of denaturation at 94℃ for 30 seconds, annealing at 62℃ for 30 seconds, extension at 72℃ for 60 seconds, and a final extension at 72℃ for 10 minutes. The amplified PCR products were visualized and analyzed on the screenTape system (Lab901 Ltd., Edinburgh, UK) with Seegene viewer software. The level of internal control/V600E amplicon band intensity was recorded by a mutation ratio >10%. In our analyses using serially diluted DNAs from the BRAF mutant cell line SNU-790, the analytical limit of detection was less than 5%.
- One hundred nanograms of DNA was amplified in a 20 µL reaction volume containing 2 µL of 10× buffer (Roche), 1.7-2.5 µM of MgCl2, 0.3 µM of each primer pair, 250 µM of dNTPs, and 2.5 unit of DNA polymerase (Roche). The PCR primers were 5'-TGCTTGCTCTGATAGGAAAATG-3' (forward) and 5'-AGCATCTCAGGGCCAAA AAT-3' (reverse). The PCR cycling conditions were as follows: an initial denaturation at 95℃ for 30 seconds, an annealing at 61℃ for 90 seconds decreasing by 0.5℃ every cycle until it reached 57℃, and an elongation step at 72℃ for 90 seconds. This procedure was followed by an additional 30 cycles with an annealing temperature at 57℃, and a final elongation step at 72℃ for 10 minutes. The PCR products were processed for the DNA sequencing reaction using the ABI-PRISM BigDye Terminator version 3.1 (Applied Biosystems) with both forward and reverse sequence-specific primers. Sequence data were generated using the ABI-PRISM 3100 DNA Analyzer (Applied Biosystems). In our analyses on the serially diluted DNA from the BRAF mutant cell line SNU-790, the analytical limit of detection was 12.5%.
- The same DNA used for DPO-PCR and direct sequencing was diluted to 5 ng/µL. A working master mix was prepared for use and 25 µL of it was added to each reaction well on the 96-well amplification-detection plates. Reaction controls and diluted specimens were then added to the respective reaction wells at 25 µL/well and mixed. The cobas 4800 BRAF V600 mutation test utilizes real-time PCR technology and uses primers that define a 116-base pair sequence of the human genomic region containing the BRAF V600E site in exon 15. BRAF wild-type and V600E target-specific fluorescent dye-labeled TaqMan probes bind to the wild-type and V600E sequences, respectively. The results were automatically generated by the cobas BRAF analysis software package (Roche Molecular Systems Inc., Branchburg, NJ, USA). In our analyses using serially diluted DNA from the BRAF mutant cell line SNU-790, the analytical limit of detection was less than 5%.
MATERIALS AND METHODS
Dual-priming oligonucleotide-PCR
Direct sequencing of the BRAF gene
Cobas 4800 BRAF V600 mutation test
- BRAF mutation results by cobas
- A total of 64 cases were finally included in this study. The summary of the test results based on the cobas test are shown in Table 1. BRAF mutations were found in tumors of 23 of the 64 patients (35.9%). In the 34 melanomas, BRAF mutations were detected in 10 cases (29.4%). Nine patients had V600E mutations and one patient harbored a V600K mutation. The frequency of BRAF mutations in malignant melanoma varied according to the primary site: 50.0% in cutaneous melanoma, 12.5% in mucosal melanoma, and 16.7% in acral melanomas. In papillary thyroid carcinomas, BRAF mutations were detected in 13 out of 16 cases (81.3%). No BRAF mutation was identified in the rest of the tumors.
- BRAF mutation concordance among dual-priming oligonucleotide-PCR, direct sequencing, and cobas
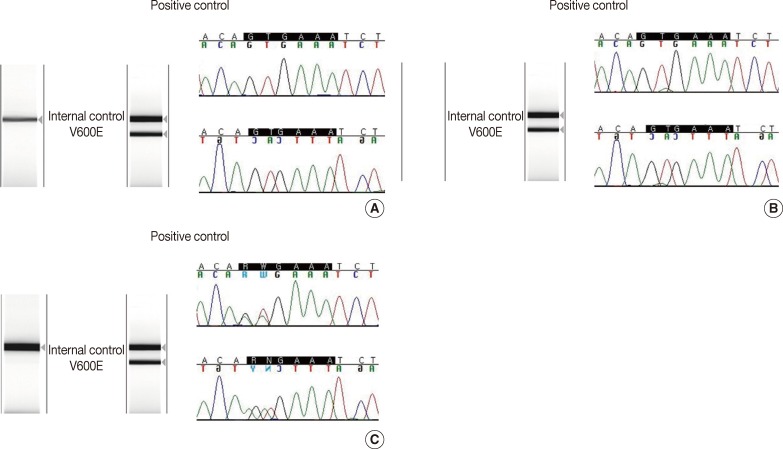
- There was 90.6% concordance (58 of 64 tested samples) and 9.4% discordance among the 3 methods. Out of the 6 discordant cases, 4 cases were melanomas and 2 cases were papillary thyroid carcinomas. The clinicopathologic characteristics of the discordant cases are provided in Table 2. Notably, 3 cases (cases nos. 27, 42, and 44) were V600E-positive only by the cobas test, and negative by DPO-PCR and direct sequencing (Fig. 1A). Among these 3 discordant cases, 1 patient (case no. 27) with a V600E-mutated metastatic melanoma which was detected only by the cobas test had been treated with vemurafenib and showed a dramatic therapeutic response. In 2 discordant cases which were confirmed to be V600E-positive in both direct sequencing and the cobas test, DPO-PCR showed invalid results with no internal control bands in repeated experiments (cases nos. 6 and 7) (Fig. 1B). In case no. 29, DPO-PCR failed to detect the V600K mutation which was identified in both direct sequencing and the cobas test (Fig. 1C). Direct sequencing revealed the V600K mutation with a substitution of 2 nucleotides (c.1798_1799 GT>AA) (Appendix 1).
RESULTS
- As the clinical efficacy of BRAF inhibitors has now been documented in BRAF V600 melanoma, the sensitivity and specificity of BRAF mutation tests to select candidate patients should be important as well. In 2011, the FDA approved vemurafenib for the treatment of metastatic melanoma patients with a BRAF V600 mutation, along with the cobas 4800 companion diagnostic test. However, how the FDA approval of the cobas 4800 diagnostic test will influence the use of the current portfolio of validated BRAF tests performed in Clinical Laboratory Improvement Amendments-certified laboratories around the country remains uncertain.15 Traditional bidirectional Sanger sequencing has been widely used for mutation testing in most clinical laboratories, but it suffers from a limited sensitivity for low-level mutant alleles and a relatively slow turn-around time.12,16
- There are scarce reports of studies which have directly compared currently available BRAF mutation tests used in the clinic.11-13,17 All previous studies have compared the analytic performance of the cobas test to Sanger sequencing and have suggested that the cobas test is superior to direct sequencing in terms of diagnostic accuracy and sensitivity. The cobas test showed a very high sensitivity (>99%) and specificity (88%) for the detection of the BRAF V600E mutation.15 In this study, BRAF mutations were tested by DPO-PCR, direct sequencing, and the cobas 4800 V600 mutation test. DPO-PCR is still performed in many laboratories in Korea because of its easy accessibility and low costs. However, the sensitivity and specificity of DPO-PCR is still controversial; The DPO system has shown results comparable to those of direct sequencing and pyrosequencing in terms of its sensitivity and specificity for detecting BRAF mutations.18 However, some have reported lower sensitivity compared to direct sequencing.19 Therefore, comparing the cobas test with DPO-PCR and direct sequencing is also important.
- An agreement analysis showed that there was discordance in 9.4% of the cases among the 3 methods. An important problem is that 4 melanomas were included in these discordant cases. According to a previous study by Halait et al.,12 the positive percent agreement between the cobas test and Sanger sequencing was 96%, and the negative percent agreement was 82%. Our study revealed 86.7% (20/23) agreement for BRAF mutation-positive cases and 100% (41/41) agreement in BRAF mutation-negative cases between the cobas test and Sanger sequencing.
- It is noteworthy that 3 cases were determined to be V600E-positive only by the cobas test and negative by DPO-PCR and direct sequencing. These three specimens contained large amounts of tumor volume and minimal known interference factors such as melanin pigments or inflammatory cells were identified. Therefore, the reason for the negative results by DPO-PCR and direct sequencing could be due to the heterogeneity of BRAF mutations existing in melanomas20,21 or the higher sensitivity of the cobas test as compared to sequencing, as was observed in previous studies.11,12 In repeated experiments, we failed to find internal positive bands in the DPO-PCR analysis. In these cases, DNA extraction was repeated several times. Although the precise cause has not been described in the literature, poor-quality DNA caused by delayed fixation or melanin pigments within tumors may have inhibited the PCR reaction.
- It is well known that Sanger sequencing is not as sensitive as mutant-specific and real-time PCR technologies. The sensitivity of direct sequencing and DPO-PCR is limited in that they are only able to detect the mutation if the tumor cells constitute more than 5% of the specimen submitted for genetic analysis.21 One concern regarding the utilization of mutation detection techniques with enhanced sensitivity is that a positive test might actually reflect the detection of a small subset of mutant cells. Some have argued that there is no clinical relevance of a tumor containing a small amount of mutant BRAF cells, as these patients would not be expected to benefit from BRAF inhibitors.21 In this study, we observed that 1 melanoma patient (case no. 27) with BRAF mutation detected only by the cobas test, but was wild-type in both DPO-PCR and direct sequencing, had been on vemurafenib treatment for 6 months and had showed a dramatic response. Further studies are therefore warranted to determine the clinical application of vemurafenib on patients with tumors containing a small amount of mutant BRAF cells or tumors with heterogeneity.
- In terms of non-V600E mutations, 1 V600K mutation was included in our study. The Catalog of Somatic Mutations in Cancer (COSMIC) database indicates that approximately 90% of BRAF codon 600 mutations in melanoma are V600E. Data are currently very limited on the clinical importance of non-V600E mutations. In the phase III trial of vemurafenib, 10 patients with V600K mutations were included and 4 of them showed partial responses, 3 had stable disease, 2 had progressive disease, and 1 had data that could not be assessed.7 These results suggest that vemurafenib may benefit patients with V600K mutations and that BRAF detection methods should be able to detect non-V600E mutation in clinical settings. Although the cobas test was designed to detect the V600E mutation, it has also showed cross-reactivity with V600K, as previously described.12
- The present study has several limitations. The sample size was small. We found only 1 V600K mutation, for which the cobas test is known to have limited detection, and the results were not confirmed by massive parallel sequencing with next-generation sequencing platforms. Given that many large cohorts report around 6-30% of melanoma tumors having V600K,22 more V600K cases are needed to better compare the methodologies.
- In conclusion, we directly compared direct sequencing with the currently available cobas test and DPO-PCR for BRAF mutations and found that the cobas test is a more sensitive method applicable for the small quantities of DNA extracted from FFPE clinical samples.
DISCUSSION
- 1. Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature 2007; 445: 851-857. ArticlePubMedPDF
- 2. Cummins DL, Cummins JM, Pantle H, Silverman MA, Leonard AL, Chanmugam A. Cutaneous malignant melanoma. Mayo Clin Proc 2006; 81: 500-507. ArticlePubMed
- 3. Cohen C, Zavala-Pompa A, Sequeira JH, et al. Mitogen-actived protein kinase activation is an early event in melanoma progression. Clin Cancer Res 2002; 8: 3728-3733. PubMed
- 4. Platz A, Egyhazi S, Ringborg U, Hansson J. Human cutaneous melanoma; a review of NRAS and BRAF mutation frequencies in relation to histogenetic subclass and body site. Mol Oncol 2008; 1: 395-405. ArticlePubMedPDF
- 5. Long GV, Menzies AM, Nagrial AM, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol 2011; 29: 1239-1246. ArticlePubMed
- 6. Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011; 364: 2507-2516. PubMedPMC
- 7. Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med 2012; 366: 707-714. ArticlePubMedPMC
- 8. Hatzivassiliou G, Song K, Yen I, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 2010; 464: 431-435. PubMed
- 9. Halaban R, Zhang W, Bacchiocchi A, et al. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells. Pigment Cell Melanoma Res 2010; 23: 190-200. PubMedPMC
- 10. Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 2010; 464: 427-430. ArticlePubMedPMCPDF
- 11. Anderson S, Bloom KJ, Vallera DU, et al. Multisite analytic performance studies of a real-time polymerase chain reaction assay for the detection of BRAF V600E mutations in formalin-fixed, paraffin-embedded tissue specimens of malignant melanoma. Arch Pathol Lab Med 2012; 136: 1385-1391. ArticlePubMedPDF
- 12. Halait H, Demartin K, Shah S, et al. Analytical performance of a real-time PCR-based assay for V600 mutations in the BRAF gene, used as the companion diagnostic test for the novel BRAF inhibitor vemurafenib in metastatic melanoma. Diagn Mol Pathol 2012; 21: 1-8. ArticlePubMed
- 13. Bloom KJ, Anderson SM, Schilling RC, et al. Molecular testing for BRAF V600 mutations in the BRIM-2 trial of the BRAF inhibitor vemurafenib in metastatic melanoma. J Clin Oncol 2011; 2(Suppl): abstr 10523.Article
- 14. Dong SM, Lee EJ, Jeon ES, Park CK, Kim KM. Progressive methylation during the serrated neoplasia pathway of the colorectum. Mod Pathol 2005; 18: 170-178. ArticlePubMedPDF
- 15. Woodman SE, Lazar AJ, Aldape KD, Davies MA. New strategies in melanoma: molecular testing in advanced disease. Clin Cancer Res 2012; 18: 1195-1200. ArticlePubMedPMCPDF
- 16. Tan YH, Liu Y, Eu KW, et al. Detection of BRAF V600E mutation by pyrosequencing. Pathology 2008; 40: 295-298. ArticlePubMed
- 17. Rueschoff J, Bloom K, Anderson S, et al. Molecular testing for BRAF V600 mutations in the phase III trial of the selective BRAF inhibitor vemurafenib in metastatic melanoma: a comparison of the cobas 4800 BRAF V600E mutation test and Sanger sequencing. Pigment Cell Melanoma Res 2011; 24: 1023.
- 18. Lee HJ, Choi J, Hwang TS, Shong YK, Hong SJ, Gong G. Detection of BRAF mutations in thyroid nodules by allele-specific PCR using a dual priming oligonucleotide system. Am J Clin Pathol 2010; 133: 802-808. ArticlePubMed
- 19. Chung KW, Yang SK, Lee GK, et al. Detection of BRAFV600E mutation on fine needle aspiration specimens of thyroid nodule refines cyto-pathology diagnosis, especially in BRAF600E mutation-prevalent area. Clin Endocrinol (Oxf) 2006; 65: 660-666. PubMed
- 20. Lin J, Goto Y, Murata H, et al. Polyclonality of BRAF mutations in primary melanoma and the selection of mutant alleles during progression. Br J Cancer 2011; 104: 464-468. ArticlePubMedPMCPDF
- 21. Sullivan RJ, Flaherty KT. BRAF in melanoma: pathogenesis, diagnosis, inhibition, and resistance. J Skin Cancer 2011; 2011: 423239.ArticlePubMedPMCPDF
- 22. Si L, Kong Y, Xu X, et al. Prevalence of BRAF V600E mutation in Chinese melanoma patients: large scale analysis of BRAF and NRAS mutations in a 432-case cohort. Eur J Cancer 2012; 48: 94-100. ArticlePubMed
REFERENCES
Appendix
Appendix 1.
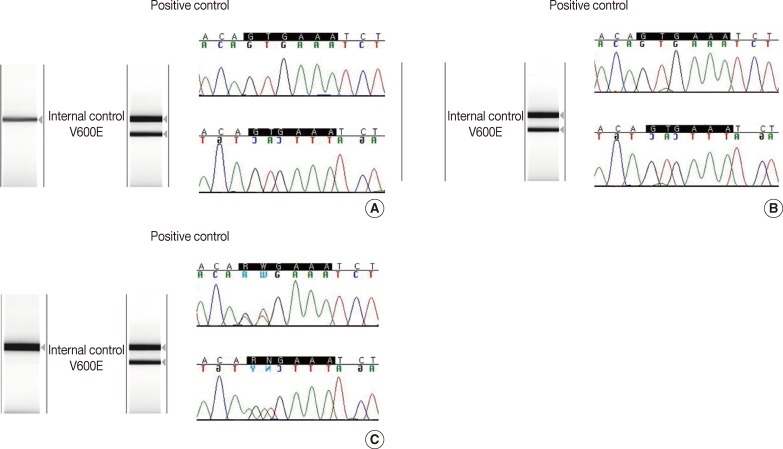
Figure & Data
References
Citations

- Preoperative BRAFV600E mutation detection in thyroid carcinoma by immunocytochemistry
Kristine Zøylner Swan, Stine Horskær Madsen, Steen Joop Bonnema, Viveque Egsgaard Nielsen, Marie Louise Jespersen
APMIS.2022; 130(11): 627. CrossRef - Strategy to reduce unnecessary surgeries in thyroid nodules with cytology of Bethesda category III (AUS/FLUS): a retrospective analysis of 667 patients diagnosed by surgery
Yong Joon Suh, Yeon Ju Choi
Endocrine.2020; 69(3): 578. CrossRef - A new primer construction technique that effectively increases amplification of rare mutant templates in samples
Jr-Kai Huang, Ling Fan, Tao-Yeuan Wang, Pao-Shu Wu
BMC Biotechnology.2019;[Epub] CrossRef - BRAF and NRAS mutations and antitumor immunity in Korean malignant melanomas and their prognostic relevance: Gene set enrichment analysis and CIBERSORT analysis
Kyueng-Whan Min, Ji-Young Choe, Mi Jung Kwon, Hye Kyung Lee, Ho Suk Kang, Eun Sook Nam, Seong Jin Cho, Hye-Rim Park, Soo Kee Min, Jinwon Seo, Yun Joong Kim, Nan Young Kim, Ho Young Kim
Pathology - Research and Practice.2019; 215(12): 152671. CrossRef - The association between dermoscopic features and BRAF mutational status in cutaneous melanoma: Significance of the blue-white veil
Miquel Armengot-Carbó, Eduardo Nagore, Zaida García-Casado, Rafael Botella-Estrada
Journal of the American Academy of Dermatology.2018; 78(5): 920. CrossRef - Comparison of Five Different Assays for the Detection of BRAF Mutations in Formalin-Fixed Paraffin Embedded Tissues of Patients with Metastatic Melanoma
Claire Franczak, Julia Salleron, Cindy Dubois, Pierre Filhine-Trésarrieu, Agnès Leroux, Jean-Louis Merlin, Alexandre Harlé
Molecular Diagnosis & Therapy.2017; 21(2): 209. CrossRef - Validation of an NGS mutation detection panel for melanoma
Anne Reiman, Hugh Kikuchi, Daniela Scocchia, Peter Smith, Yee Wah Tsang, David Snead, Ian A Cree
BMC Cancer.2017;[Epub] CrossRef - Transformation to Small Cell Lung Cancer of Pulmonary Adenocarcinoma: Clinicopathologic Analysis of Six Cases
Soomin Ahn, Soo Hyun Hwang, Joungho Han, Yoon-La Choi, Se-Hoon Lee, Jin Seok Ahn, Keunchil Park, Myung-Ju Ahn, Woong-Yang Park
Journal of Pathology and Translational Medicine.2016; 50(4): 258. CrossRef - Immunohistochemistry with the anti-BRAF V600E (VE1) antibody: impact of pre-analytical conditions and concordance with DNA sequencing in colorectal and papillary thyroid carcinoma
Katerina Dvorak, Birte Aggeler, John Palting, Penny McKelvie, Andrew Ruszkiewicz, Paul Waring
Pathology.2014; 46(6): 509. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure

Fig. 1
| Case No. | Pathologic diagnosis | DPO-PCR | Sequencing | cobas | Case No. | Pathologic diagnosis | DPO-PCR | Sequencing | cobas | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Malignant melanoma | Wild | Wild | Wild | 33 | Malignant melanoma | Mutant (V600E) | Mutant (V600E) | Mutant | |
| 2 | Malignant melanoma | Wild | Wild | Wild | 34 | Malignant melanoma | Wild | Wild | Wild | |
| 3 | Malignant melanoma | Mutant (V600E) | Mutant (V600E) | Mutant | 35 | Papillary carcinoma | Mutant (V600E) | Mutant (V600E) | Mutant | |
| 4 | Malignant melanoma | Mutant (V600E) | Mutant (V600E) | Mutant | 36 | Papillary carcinoma | Wild | Wild | Wild | |
| 5 | Malignant melanoma | Wild | Wild | Wild | 37 | Papillary carcinoma | Mutant (V600E) | Mutant (V600E) | Mutant | |
| 6 | Malignant melanoma | Failed | Mutant (V600E) | Mutant | 38 | Papillary carcinoma | Mutant (V600E) | Mutant (V600E) | Mutant | |
| 7 | Malignant melanoma | Failed | Mutant (V600E) | Mutant | 39 | Papillary carcinoma | Wild | Wild | Wild | |
| 8 | Malignant melanoma | Mutant (V600E) | Mutant (V600E) | Mutant | 40 | Papillary carcinoma | Wild | Wild | Wild | |
| 9 | Malignant melanoma | Wild | Wild | Wild | 41 | Papillary carcinoma | Mutant (V600E) | Mutant (V600E) | Mutant | |
| 10 | Malignant melanoma | Wild | Wild | Wild | 42 | Papillary carcinoma | Wild | Wild | Mutant | |
| 11 | Malignant melanoma | Wild | Wild | Wild | 43 | Papillary carcinoma | Mutant (V600E) | Mutant (V600E) | Mutant | |
| 12 | Malignant melanoma | Wild | Wild | Wild | 44 | Papillary carcinoma | Wild | Wild | Mutant | |
| 13 | Malignant melanoma | Wild | Wild | Wild | 45 | Papillary carcinoma | Mutant (V600E) | Mutant (V600E) | Mutant | |
| 14 | Malignant melanoma | Wild | Wild | Wild | 46 | Papillary carcinoma | Mutant (V600E) | Mutant (V600E) | Mutant | |
| 15 | Malignant melanoma | Wild | Wild | Wild | 47 | Papillary carcinoma | Mutant (V600E) | Mutant (V600E) | Mutant | |
| 16 | Malignant melanoma | Wild | Wild | Wild | 48 | Papillary carcinoma | Mutant (V600E) | Mutant (V600E) | Mutant | |
| 17 | Malignant melanoma | Mutant (V600E) | Mutant (V600E) | Mutant | 49 | Papillary carcinoma | Mutant (V600E) | Mutant (V600E) | Mutant | |
| 18 | Malignant melanoma | Wild | Wild | Wild | 50 | Papillary carcinoma | Mutant (V600E) | Mutant (V600E) | Mutant | |
| 19 | Malignant melanoma | Wild | Wild | Wild | 51 | Adenocarcinoma | Wild | Wild | Wild | |
| 20 | Malignant melanoma | Wild | Wild | Wild | 52 | Adenocarcinoma | Wild | Wild | Wild | |
| 21 | Malignant melanoma | Wild | Wild | Wild | 53 | Adenocarcinoma | Wild | Wild | Wild | |
| 22 | Malignant melanoma | Wild | Wild | Wild | 54 | Adenocarcinoma | Wild | Wild | Wild | |
| 23 | Malignant melanoma | Wild | Wild | Wild | 55 | Adenocarcinoma | Wild | Wild | Wild | |
| 24 | Malignant melanoma | Wild | Wild | Wild | 56 | Adenocarcinoma | Wild | Wild | Wild | |
| 25 | Malignant melanoma | Wild | Wild | Wild | 57 | Adenocarcinoma | Wild | Wild | Wild | |
| 26 | Malignant melanoma | Mutant (V600E) | Mutant (V600E) | Mutant | 58 | Adenocarcinoma | Wild | Wild | Wild | |
| 27 | Malignant melanoma | Wild | Wild | Mutant | 59 | Adenocarcinoma | Wild | Wild | Wild | |
| 28 | Malignant melanoma | Wild | Wild | Wild | 60 | Adenocarcinoma | Wild | Wild | Wild | |
| 29 | Malignant melanoma | Wild | Mutant (V600K) | Mutant | 61 | Adenocarcinoma | Wild | Wild | Wild | |
| 30 | Malignant melanoma | Wild | Wild | Wild | 62 | Clear cell carcinoma | Wild | Wild | Wild | |
| 31 | Malignant melanoma | Wild | Wild | Wild | 63 | Mucinous adenocarcinoma | Wild | Wild | Wild | |
| 32 | Malignant melanoma | Wild | Wild | Wild | 64 | Epithelioid sarcoma | Wild | Wild | Wild |
| Pathologic diagnosis and origin of tumor | Case No. | Tumors with BRAF mutation |
|||
|---|---|---|---|---|---|
| Wild | V600E | V600K | |||
| Malignant melanoma | |||||
| Primary | Cutaneous | 12 | 6 (50.0) | 5 (41.7) | 1 (8.3) |
| Mucosal | 8 | 7 (87.5) | 1 (12.5) | 0 | |
| Acral | 12 | 10 (83.3) | 2 (16.7) | 0 | |
| Unknown | 2 | 1 (50) | 1 (50) | 0 | |
| Total | 34 | 24 (70.6) | 9 (26.5) | 1 (2.9) | |
| Papillary carcinoma from thyroid | 16 | 3 (18.8) | 13 (81.2) | 0 | |
| Adenocarcinoma from colon | 11 | 11 (100) | 0 | 0 | |
| Clear cell carcinoma from skin | 1 | 1 (100) | 0 | 0 | |
| Mucinous adenocarcinoma from bladder | 1 | 1 (100) | 0 | 0 | |
| Epithelioid sarcoma | 1 | 1 (100) | 0 | 0 | |
| Case No. | Tumor type | DPO-PCR | Sequencing | cobas | Site of organ obtained | Tumor volume (%) | Melanin pigments | Necrosis (%) | Lymphocytes content (%) | DNA concentration (ng/μL) | 260/280 ratio |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | Malignant melanoma | Failed | Mutant (V600E) | Mutant | Skin (thumb) | 70 | Minimal | 0 | 5 | 42.7 | 1.53 |
| 7 | Malignant melanoma | Failed | Mutant (V600E) | Mutant | Skin (scalp) | 95 | Minimal | 0 | < 5 | 8.3 | 0.88 |
| 27 | Malignant melanoma | Wild | Wild | Mutant | Lymph node | 90 | Minimal | 4 | 5 | 35.8 | 1.49 |
| 29 | Malignant melanoma | Wild | Mutant (V600K) | Mutant | Lymph node | 50 | Moderate | 2 | 50 | 26.5 | 1.45 |
| 42 | Papillary carcinoma | Wild | Wild | Mutant | Thyroid | 40 | No | 0 | < 5 | 52.3 | 1.95 |
| 44 | Papillary carcinoma | Wild | Wild | Mutant | Thyroid | 80 | No | 0 | < 5 | 60.8 | 1.98 |
Bold are cases with discordant results.
Values are presented as number (%).

 E-submission
E-submission