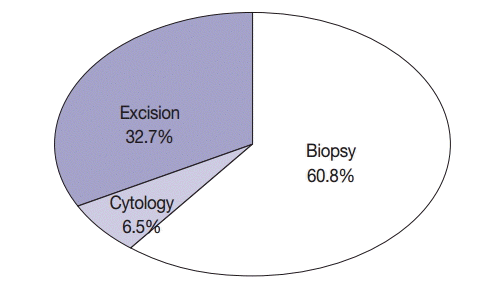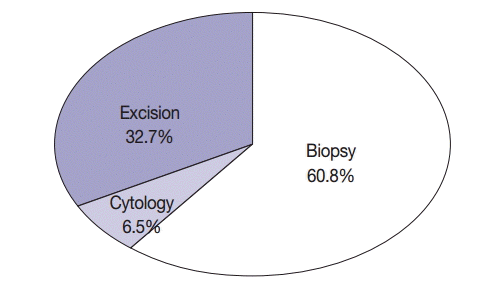Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 49(6); 2015 > Article
-
Original Article
Analysis of Mutations in Epidermal Growth Factor Receptor Gene in Korean Patients with Non-small Cell Lung Cancer: Summary of a Nationwide Survey - Sang Hwa Lee, Wan Seop Kim, Yoo Duk Choi1, Jeong Wook Seo2, Joung Ho Han3, Mi Jin Kim4, Lucia Kim5, Geon Kook Lee6, Chang Hun Lee7, Mee Hye Oh8, Gou Young Kim9, Sun Hee Sung10, Kyo Young Lee11, Sun Hee Chang12, Mee Sook Rho13, Han Kyeom Kim14, Soon Hee Jung15, Se Jin Jang16, The Cardiopulmonary Pathology Study Group of Korean Society of Pathologists
-
Journal of Pathology and Translational Medicine 2015;49(6):481-488.
DOI: https://doi.org/10.4132/jptm.2015.09.14
Published online: October 13, 2015
Department of Pathology, Konkuk University School of Medicine, Seoul, Korea
1Chonnam National University Medical School, Gwangju, Korea
2Seoul National University College of Medicine, Seoul, Korea
3Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
4Yeungnam University College of Medicine, Daegu, Korea
5Inha University College of Medicine, Incheon, Korea
6National Cancer Center, Goyang, Korea
7Busan National University School of Medicine, Busan, Korea
8Soonchunhyang University Cheonan Hospital, Soonchunhyang University College of Medicine, Cheonan, Korea
9Kyung Hee University School of Medicine, Seoul, Korea
10Ewha Womans University School of Medicine, Seoul, Korea
11Seoul St. Mary’s Hospital, Catholic University of Korea, Seoul, Korea
12Inje University Ilsan Paik Hospital, Goyang, Korea
13Dong-A University College of Medicine, Busan, Korea
14Korea University College of Medicine, Seoul, Korea
15Yonsei University Wonju College of Medicine, Wonju, Korea
16Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- Corresponding Author: Wan Seop Kim, MD, PhD Department of Pathology, Konkuk University Medical Center, Konkuk University School of Medicine, 120-1 Neungdong-ro, Gwangjin-gu, Seoul 05030, Korea Tel: +82-2-2030-5642 Fax: +82-2-2030-5629 E-mail: wskim@kuh.ac.kr
© 2015 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Figure & Data
References
Citations

- Assessing mutation-clinical correlations and treatment outcomes in Vietnamese non-small cell lung cancer patients
Hoang-Bac Nguyen, Bang-Suong Nguyen-Thi, Huu-Huy Nguyen, Minh-Khoi Le, Quoc-Trung Lam, Tuan-Anh Nguyen
Practical Laboratory Medicine.2025; 45: e00477. CrossRef - Projected cancer burden attributable to population aging: Insight from a rapidly aging society
Minh‐Thao Tu, Hoejun Kwon, Yoon‐Jung Choi, Hyunsoon Cho
International Journal of Cancer.2025;[Epub] CrossRef - Gradual Increase in Lung Cancer Risk Due to Particulate Matter Exposure in Patients With Pulmonary Function Impairments: A Nationwide Korean Database Analysis
Jongin Lee, Joon Young Choi, Jeong Uk Lim
Journal of Korean Medical Science.2025;[Epub] CrossRef - The role of oncogenes and tumor suppressor genes in determining survival rates of lung cancer patients in the population of North Sumatra, Indonesia
Noni Novisari Soeroso, Fannie Rizki Ananda, Johan Samuel Sitanggang, Noverita Sprinse Vinolina
F1000Research.2023; 11: 853. CrossRef - Comprehensive analysis of NGS and ARMS-PCR for detecting EGFR mutations based on 4467 cases of NSCLC patients
Changlong He, Chengcheng Wei, Jun Wen, Shi Chen, Ling Chen, Yue Wu, Yifan Shen, Huili Bai, Yangli Zhang, Xueping Chen, Xiaosong Li
Journal of Cancer Research and Clinical Oncology.2022; 148(2): 321. CrossRef - Unique characteristics of G719X and S768I compound double mutations of epidermal growth factor receptor (EGFR) gene in lung cancer of coal-producing areas of East Yunnan in Southwestern China
Jun-Ling Wang, Yu-Dong Fu, Yan-Hong Gao, Xiu-Ping Li, Qian Xiong, Rui Li, Bo Hou, Ruo-Shan Huang, Jun-Feng Wang, Jian-Kun Zhang, Jia-Ling Lv, Chao Zhang, Hong-Wei Li
Genes and Environment.2022;[Epub] CrossRef - Continuous Vaginal Bleeding Induced By EGFR-TKI in Premenopausal Female Patients With EGFR Mutant NSCLC
Min Yu, Xiaoyu Li, Xueqian Wu, Weiya Wang, Yanying Li, Yan Zhang, Shuang Zhang, Yongsheng Wang
Frontiers in Oncology.2022;[Epub] CrossRef - The role of oncogenes and tumor suppressor genes in determining survival rates of lung cancer patients in the population of North Sumatra, Indonesia
Noni Novisari Soeroso, Fannie Rizki Ananda, Johan Samuel Sitanggang, Noverita Sprinse Vinolina
F1000Research.2022; 11: 853. CrossRef - Adverse Event Profiles of Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Adenocarcinoma Lung Patients in North Sumatera Population
Moh. Ramadhani Soeroso, Noni Novisari Soeroso, Setia Putra Tarigan, Elisna Syahruddin
Open Access Macedonian Journal of Medical Sciences.2022; 10(T7): 134. CrossRef - Landscape of EGFR mutations in lung adenocarcinoma: a single institute experience with comparison of PANAMutyper testing and targeted next-generation sequencing
Jeonghyo Lee, Yeon Bi Han, Hyun Jung Kwon, Song Kook Lee, Hyojin Kim, Jin-Haeng Chung
Journal of Pathology and Translational Medicine.2022; 56(5): 249. CrossRef - Traditional Chinese Medicine Syndromes are Associated with Driver Gene Mutations and Clinical Characteristics in Patients with Lung Adenocarcinoma
Jili Yang, Haiyan Lu, Niancai Jing, Bo Wang, Huanyu Guo, Shoukun Sun, Yue Zhang, Chan-Yen Kuo
Evidence-Based Complementary and Alternative Medicine.2022; 2022: 1. CrossRef - Exosome-based detection of EGFR T790M in plasma and pleural fluid of prospectively enrolled non-small cell lung cancer patients after first-line tyrosine kinase inhibitor therapy
Yoonjung Kim, Saeam Shin, Kyung-A Lee
Cancer Cell International.2021;[Epub] CrossRef - Molecular biomarker testing for non–small cell lung cancer: consensus statement of the Korean Cardiopulmonary Pathology Study Group
Sunhee Chang, Hyo Sup Shim, Tae Jung Kim, Yoon-La Choi, Wan Seop Kim, Dong Hoon Shin, Lucia Kim, Heae Surng Park, Geon Kook Lee, Chang Hun Lee
Journal of Pathology and Translational Medicine.2021; 55(3): 181. CrossRef - Osimertinib in Patients with T790M-Positive Advanced Non-small Cell Lung Cancer: Korean Subgroup Analysis from Phase II Studies
Myung-Ju Ahn, Ji-Youn Han, Dong-Wan Kim, Byoung Chul Cho, Jin-Hyoung Kang, Sang-We Kim, James Chih-Hsin Yang, Tetsuya Mitsudomi, Jong Seok Lee
Cancer Research and Treatment.2020; 52(1): 284. CrossRef - Long non-coding RNA ATB promotes human non-small cell lung cancer proliferation and metastasis by suppressing miR-141-3p
Guojie Lu, Yaosen Zhang, Klaus Roemer
PLOS ONE.2020; 15(2): e0229118. CrossRef - Prognostic Role of S100A8 and S100A9 Protein Expressions in Non-small Cell Carcinoma of the Lung
Hyun Min Koh, Hyo Jung An, Gyung Hyuck Ko, Jeong Hee Lee, Jong Sil Lee, Dong Chul Kim, Jung Wook Yang, Min Hye Kim, Sung Hwan Kim, Kyung Nyeo Jeon, Gyeong-Won Lee, Se Min Jang, Dae Hyun Song
Journal of Pathology and Translational Medicine.2019; 53(1): 13. CrossRef - Epidermal growth factor receptor T790M mutations in non-small cell lung cancer (NSCLC) of Yunnan in southwestern China
Yongchun Zhou, Yuhui Ma, Hutao Shi, Yaxi Du, Yunchao Huang
Scientific Reports.2018;[Epub] CrossRef - Does the efficacy of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor differ according to the type of EGFR mutation in non-small cell lung cancer?
Yong Won Choi, Jin-Hyuk Choi
The Korean Journal of Internal Medicine.2017; 32(3): 422. CrossRef - Molecular Testing of Lung Cancers
Hyo Sup Shim, Yoon-La Choi, Lucia Kim, Sunhee Chang, Wan-Seop Kim, Mee Sook Roh, Tae-Jung Kim, Seung Yeon Ha, Jin-Haeng Chung, Se Jin Jang, Geon Kook Lee
Journal of Pathology and Translational Medicine.2017; 51(3): 242. CrossRef - MET Exon 14 Skipping Mutations in Lung Adenocarcinoma: Clinicopathologic Implications and Prognostic Values
Geun Dong Lee, Seung Eun Lee, Doo-Yi Oh, Dan-bi Yu, Hae Min Jeong, Jooseok Kim, Sungyoul Hong, Hun Soon Jung, Ensel Oh, Ji-Young Song, Mi-Sook Lee, Mingi Kim, Kyungsoo Jung, Jhingook Kim, Young Kee Shin, Yoon-La Choi, Hyeong Ryul Kim
Journal of Thoracic Oncology.2017; 12(8): 1233. CrossRef - Epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer (NSCLC) of Yunnan in southwestern China
Yongchun Zhou, Yanlong Yang, Chenggang Yang, Yunlan Chen, Changshao Yang, Yaxi Du, Guangqiang Zhao, Yinjin Guo, Lianhua Ye, Yunchao Huang
Oncotarget.2017; 8(9): 15023. CrossRef - Detection of EGFR and KRAS Mutation by Pyrosequencing Analysis in Cytologic Samples of Non-Small Cell Lung Cancer
Seung Eun Lee, So-Young Lee, Hyung-Kyu Park, Seo-Young Oh, Hee-Joung Kim, Kye-Young Lee, Wan-Seop Kim
Journal of Korean Medical Science.2016; 31(8): 1224. CrossRef - MassARRAY, pyrosequencing, and PNA clamping for EGFR mutation detection in lung cancer tissue and cytological samples: a multicenter study
Kyueng-Whan Min, Wan-Seop Kim, Se Jin Jang, Yoo Duk Choi, Sunhee Chang, Soon Hee Jung, Lucia Kim, Mee-Sook Roh, Choong Sik Lee, Jung Weon Shim, Mi Jin Kim, Geon Kook Lee
Journal of Cancer Research and Clinical Oncology.2016; 142(10): 2209. CrossRef - Clinicopathologic characteristics of EGFR, KRAS, and ALK alterations in 6,595 lung cancers
Boram Lee, Taebum Lee, Se-Hoon Lee, Yoon-La Choi, Joungho Han
Oncotarget.2016; 7(17): 23874. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure




Fig. 1.
Fig. 2.
Fig. 3.
Fig. 4.
| Characteristic | No. (%) |
|---|---|
| Age, mean (range, yr) | 62.7 (16–89) |
| < 65 | 878 (50.1) |
| ≥ 65 | 875 (49.9) |
| Sex | |
| Female | 753 (43) |
| Male | 1,000 (57) |
| Smoking history | |
| Smoker | 555 (31.7) |
| Light smoker | 39 (2.2) |
| Ex-smoker | 170 (9.7) |
| Never smoker | 849 (48.4) |
| Unknown | 140 (8.0) |
| Specimen type | |
| Cytology | 114 (6.5) |
| Biopsy | 1,066 (60.8) |
| Excision | 573 (32.7) |
| Procurement site | |
| Primary | 1,544 (88.1) |
| Metastasis | 209 (11.9) |
| Tumor dissection | |
| No | 741 (42.3) |
| Yes | 1,012 (57.7) |
| Histologic type | |
| Adenocarcinoma | 1,292 (73.7) |
| Squamous cell carcinoma | 347 (19.8) |
| Non-small cell carcinoma | 69 (3.9) |
| Pleomorphic carcinoma | 13 (0.7) |
| Large cell neuroendocrine carcinoma | 12 (0.7) |
| Large cell carcinoma | 9 (0.5) |
| Sarcomatoid carcinoma | 5 (0.3) |
| Carcinoid tumor | 2 (0.1) |
| Mucoepidermoid carcinoma | 2 (0.1) |
| Carcinosarcoma | 1 (0.05) |
| Lymphoepithelioma-like carcinoma | 1 (0.05) |
| Laboratory status | |
| Out-sourcing | 454 (25.9) |
| In-house | 1,299 (74.1) |
| Variable | NSCLC |
Adenocarcinoma |
SqCC |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | p-value | Negative | Positive | p-value | Negative | Positive | p-value | |
| Sex | < .001 | < .001 | .001 | ||||||
| Female | 374 (49.7) | 379 (50.3) | 314 (46.5) | 361 (53.5) | 31 (75.6) | 10 (24.4) | |||
| Male | 778 (77.8) | 222 (22.2) | 418 (67.7) | 199 (32.3) | 286 (93.5) | 20 (6.5) | |||
| Age (yr) | .007 | .911 | .565 | ||||||
| < 65 | 551 (62.7) | 328 (37.3) | 399 (56.5) | 307 (43.5) | 100 (90.1) | 11 (9.9) | |||
| > 65 | 601 (68.8) | 273 (31.2) | 333 (56.8) | 253 (43.2) | 217 (91.9) | 19 (8.1) | |||
| Smoking | < .001 | < .001 | .017 | ||||||
| Ex-smoker | 143 (84.1) | 27 (15.9) | 88 (77.2) | 26 (22.8) | 48 (98) | 1 (2) | |||
| Light smoker | 22 (56.4) | 17 (43.6) | 17 (53.1) | 15 (46.9) | 2 (66.7) | 1 (33.3) | |||
| Never smoker | 442 (52.1) | 407 (47.9) | 353 (47.6) | 389 (52.4) | 58 (82.9) | 12 (17.1) | |||
| Smoker | 446 (80.4) | 109 (19.6) | 227 (70.7) | 94 (29.3) | 168 (92.8) | 13 (7.2) | |||
| Unknown | 99 (70.7) | 41 (29.3) | 47 (56.6) | 36 (43.4) | 41 (93.2) | 3 (6.8) | |||
| Specimen type | .002 | < .001 | .535 | ||||||
| Biopsy | 728 (68.3) | 338 (31.7) | 453 (59.8) | 305 (40.2) | 215 (90.7) | 22 (9.3) | |||
| Cytology | 80 (70.2) | 34 (29.8) | 70 (67.3) | 34 (32.6) | |||||
| Excision | 344 (60) | 229 (40) | 209 (48.6) | 221 (51.4) | 102 (92.7) | 8 (7.3) | |||
| Procurement site | .797 | .586 | .38 | ||||||
| Metastasis | 139 (66.5) | 70 (33.5) | 95 (58.6) | 67 (41.4) | 17 (100) | 0 (0) | |||
| Primary | 1,013 (65.6) | 531 (34.4) | 637 (56.4) | 493 (43.6) | 300 (90.9) | 30 (9.1) | |||
| Tumor dissection | .362 | .001 | .105 | ||||||
| No | 478 (64.5) | 263 (35.5) | 401 (61.3) | 253 (38.7) | 45 (84.9) | 8 (15.1) | |||
| Yes | 674 (66.6) | 338 (33.4) | 331 (51.9) | 307 (48.1) | 272 (92.5) | 22 (7.5) | |||
| Laboratory status | < .001 | .046 | .238 | ||||||
| In-house | 803 (61.8) | 496 (38.2) | 581 (55.3) | 469 (44.7) | 144 (89.4) | 17 (10.6) | |||
| Out-sourcing | 349 (76.9) | 105 (23.1) | 151 (62.4) | 91 (37.6) | 173 (93) | 13 (7) | |||
Values are presented as number (%).

 E-submission
E-submission