Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 51(3); 2017 > Article
-
Review
Good Laboratory Standards for Clinical Next-Generation Sequencing Cancer Panel Tests - Jihun Kim1,2,*, Woong-Yang Park3,*, Nayoung K. D. Kim3, Se Jin Jang1,2, Sung-Min Chun1,2, Chang-Ohk Sung1,2, Jene Choi1, Young-Hyeh Ko4, Yoon-La Choi4, Hyo Sup Shim5, Jae-Kyung Won6, The Molecular Pathology Study Group of Korean Society of Pathologists
-
Journal of Pathology and Translational Medicine 2017;51(3):191-204.
DOI: https://doi.org/10.4132/jptm.2017.03.14
Published online: May 10, 2017
1Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
2Center for Cancer Genome Discovery, Asan Institute for Life Sciences, Seoul, , Korea
3Samsung Genome Institute, Sungkyunkwan University School of Medicine, Seoul, Korea
4Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
5Department of Pathology, Yonsei University College of Medicine, Seoul, Korea
6Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
- Corresponding Author Jihun Kim, MD, PhD Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, 88 Olympic-ro 43-gil, Songpa-gu, Seoul 05505, Korea Tel: +82-2-3010-4556 Fax: +82-2-472-7898 E-mail: jihunkim@amc.seoul.kr
- *Jihun Kim and Woong-Yang Park contributed equally to this work as co-first authors.
© 2017 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Figure & Data
References
Citations

- Tumour purity assessment with deep learning in colorectal cancer and impact on molecular analysis
Lydia A Schoenpflug, Aikaterini Chatzipli, Korsuk Sirinukunwattana, Susan Richman, Andrew Blake, James Robineau, Kirsten D Mertz, Clare Verrill, Simon J Leedham, Claire Hardy, Celina Whalley, Keara Redmond, Philip Dunne, Steven Walker, Andrew D Beggs, Ult
The Journal of Pathology.2025; 265(2): 184. CrossRef - Clinical Validation of Local Versus Commercial Genomic Testing in Cancer: A Comparison of Tissue and Plasma Concordance
Lucy G. Faulkner, Lynne Howells, Susann Lehman, Caroline Cowley, Zahirah Sidat, Jacqui Shaw, Anne L. Thomas
Cancer Investigation.2025; 43(2): 119. CrossRef - Diagnostic Implications of NGS-Based Molecular Profiling in Mature B-Cell Lymphomas with Potential Bone Marrow Involvement
Bernhard Strasser, Sebastian Mustafa, Josef Seier, Erich Wimmer, Josef Tomasits
Diagnostics.2025; 15(6): 727. CrossRef - Pragmatic nationwide master observational trial based on genomic alterations in advanced solid tumors: KOrean Precision Medicine Networking Group Study of MOlecular profiling guided therapy based on genomic alterations in advanced Solid tumors (KOSMOS)-II
Sun Young Kim, Jee Hyun Kim, Tae-Yong Kim, Sook Ryun Park, Shinkyo Yoon, Soohyeon Lee, Se-Hoon Lee, Tae Min Kim, Sae-Won Han, Hye Ryun Kim, Hongseok Yun, Sejoon Lee, Jihun Kim, Yoon-La Choi, Kui Son Choi, Heejung Chae, Hyewon Ryu, Gyeong-Won Lee, Dae Youn
BMC Cancer.2024;[Epub] CrossRef - Reporting of somatic variants in clinical cancer care: recommendations of the Swiss Society of Molecular Pathology
Yann Christinat, Baptiste Hamelin, Ilaria Alborelli, Paolo Angelino, Valérie Barbié, Bettina Bisig, Heather Dawson, Milo Frattini, Tobias Grob, Wolfram Jochum, Ronny Nienhold, Thomas McKee, Matthias Matter, Edoardo Missiaglia, Francesca Molinari, Sacha Ro
Virchows Archiv.2024; 485(6): 1033. CrossRef - Acute myeloid leukemia and myelodysplastic neoplasms: clinical implications of myelodysplasia-related genes mutations and TP53 aberrations
Hyunwoo Kim, Ja Young Lee, Shinae Yu, Eunkyoung Yoo, Hye Ran Kim, Sang Min Lee, Won Sik Lee
Blood Research.2024;[Epub] CrossRef - Validation and Clinical Application of ONCOaccuPanel for Targeted Next-Generation Sequencing of Solid Tumors
Moonsik Kim, Changseon Lee, Juyeon Hong, Juhee Kim, Ji Yun Jeong, Nora Jee-Young Park, Ji-Eun Kim, Ji Young Park
Cancer Research and Treatment.2023; 55(2): 429. CrossRef - Establishing molecular pathology curriculum for pathology trainees and continued medical education: a collaborative work from the Molecular Pathology Study Group of the Korean Society of Pathologists
Jiwon Koh, Ha Young Park, Jeong Mo Bae, Jun Kang, Uiju Cho, Seung Eun Lee, Haeyoun Kang, Min Eui Hong, Jae Kyung Won, Youn-La Choi, Wan-Seop Kim, Ahwon Lee
Journal of Pathology and Translational Medicine.2023; 57(5): 265. CrossRef - Clinical applications of next-generation sequencing in the diagnosis of genetic disorders in Korea: a narrative review
Jihoon G. Yoon, Man Jin Kim, Yong Jin Kwon, Jong-Hee Chae
Journal of the Korean Medical Association.2023; 66(10): 613. CrossRef - Obtaining spatially resolved tumor purity maps using deep multiple instance learning in a pan-cancer study
Mustafa Umit Oner, Jianbin Chen, Egor Revkov, Anne James, Seow Ye Heng, Arife Neslihan Kaya, Jacob Josiah Santiago Alvarez, Angela Takano, Xin Min Cheng, Tony Kiat Hon Lim, Daniel Shao Weng Tan, Weiwei Zhai, Anders Jacobsen Skanderup, Wing-Kin Sung, Hwee
Patterns.2022; 3(2): 100399. CrossRef - Update on Molecular Diagnosis in Extranodal NK/T-Cell Lymphoma and Its Role in the Era of Personalized Medicine
Ka-Hei (Murphy) Sun, Yin-Ting (Heylie) Wong, Ka-Man (Carmen) Cheung, Carmen (Michelle) Yuen, Yun-Tat (Ted) Chan, Wing-Yan (Jennifer) Lai, Chun (David) Chao, Wing-Sum (Katie) Fan, Yuen-Kiu (Karen) Chow, Man-Fai Law, Ho-Chi (Tommy) Tam
Diagnostics.2022; 12(2): 409. CrossRef - Defining Novel DNA Virus-Tumor Associations and Genomic Correlates Using Prospective Clinical Tumor/Normal Matched Sequencing Data
Chad M. Vanderbilt, Anita S. Bowman, Sumit Middha, Kseniya Petrova-Drus, Yi-Wei Tang, Xin Chen, Youxiang Wang, Jason Chang, Natasha Rekhtman, Klaus J. Busam, Sounak Gupta, Meera Hameed, Maria E. Arcila, Marc Ladanyi, Michael F. Berger, Snjezana Dogan, Ahm
The Journal of Molecular Diagnostics.2022; 24(5): 515. CrossRef - Performance Evaluation of Three DNA Sample Tracking Tools in a Whole Exome Sequencing Workflow
Gertjan Wils, Céline Helsmoortel, Pieter-Jan Volders, Inge Vereecke, Mauro Milazzo, Jo Vandesompele, Frauke Coppieters, Kim De Leeneer, Steve Lefever
Molecular Diagnosis & Therapy.2022; 26(4): 411. CrossRef - Clinical Quality Considerations when Using Next-Generation Sequencing (NGS) in Clinical Drug Development
Timothé Ménard, Alaina Barros, Christopher Ganter
Therapeutic Innovation & Regulatory Science.2021; 55(5): 1066. CrossRef - Fast Healthcare Interoperability Resources (FHIR)–Based Quality Information Exchange for Clinical Next-Generation Sequencing Genomic Testing: Implementation Study
Donghyeong Seong, Sungwon Jung, Sungchul Bae, Jongsuk Chung, Dae-Soon Son, Byoung-Kee Yi
Journal of Medical Internet Research.2021; 23(4): e26261. CrossRef - Status of Next-Generation Sequencing-Based Genetic Diagnosis in Hematologic Malignancies in Korea (2017-2018)
JinJu Kim, Ja Young Lee, Jungwon Huh, Myung-Hyun Nam, Myungshin Kim, Young-Uk Cho, Sun-Young Kong, Seung-Tae Lee, In-Suk Kim
Laboratory Medicine Online.2021; 11(1): 25. CrossRef - MSI-Testung
Josef Rüschoff, Gustavo Baretton, Hendrik Bläker, Wolfgang Dietmaier, Manfred Dietel, Arndt Hartmann, Lars-Christian Horn, Korinna Jöhrens, Thomas Kirchner, Ruth Knüchel, Doris Mayr, Sabine Merkelbach-Bruse, Hans-Ulrich Schildhaus, Peter Schirmacher, Mark
Der Pathologe.2021; 42(4): 414. CrossRef - Molecular biomarker testing for non–small cell lung cancer: consensus statement of the Korean Cardiopulmonary Pathology Study Group
Sunhee Chang, Hyo Sup Shim, Tae Jung Kim, Yoon-La Choi, Wan Seop Kim, Dong Hoon Shin, Lucia Kim, Heae Surng Park, Geon Kook Lee, Chang Hun Lee
Journal of Pathology and Translational Medicine.2021; 55(3): 181. CrossRef - MSI testing
Josef Rüschoff, Gustavo Baretton, Hendrik Bläker, Wolfgang Dietmaier, Manfred Dietel, Arndt Hartmann, Lars-Christian Horn, Korinna Jöhrens, Thomas Kirchner, Ruth Knüchel, Doris Mayr, Sabine Merkelbach-Bruse, Hans-Ulrich Schildhaus, Peter Schirmacher, Mark
Der Pathologe.2021; 42(S1): 110. CrossRef - 16S rDNA microbiome composition pattern analysis as a diagnostic biomarker for biliary tract cancer
Huisong Lee, Hyeon Kook Lee, Seog Ki Min, Won Hee Lee
World Journal of Surgical Oncology.2020;[Epub] CrossRef - Risk Stratification Using a Novel Genetic Classifier IncludingPLEKHS1Promoter Mutations for Differentiated Thyroid Cancer with Distant Metastasis
Chan Kwon Jung, Seung-Hyun Jung, Sora Jeon, Young Mun Jeong, Yourha Kim, Sohee Lee, Ja-Seong Bae, Yeun-Jun Chung
Thyroid.2020; 30(11): 1589. CrossRef - Biomarker testing for advanced lung cancer by next-generation sequencing; a valid method to achieve a comprehensive glimpse at mutational landscape
Anurag Mehta, Smreti Vasudevan, Sanjeev Kumar Sharma, Manoj Panigrahi, Moushumi Suryavanshi, Mumtaz Saifi, Ullas Batra
Applied Cancer Research.2020;[Epub] CrossRef - Application Areas of Traditional Molecular Genetic Methods and NGS in relation to Hereditary Urological Cancer Diagnosis
Dmitry S. Mikhaylenko, Alexander S. Tanas, Dmitry V. Zaletaev, Marina V. Nemtsova
Journal of Oncology.2020; 2020: 1. CrossRef - Assembling and Validating Bioinformatic Pipelines for Next-Generation Sequencing Clinical Assays
Jeffrey A SoRelle, Megan Wachsmann, Brandi L. Cantarel
Archives of Pathology & Laboratory Medicine.2020; 144(9): 1118. CrossRef - Standard operating procedure for somatic variant refinement of sequencing data with paired tumor and normal samples
Erica K. Barnell, Peter Ronning, Katie M. Campbell, Kilannin Krysiak, Benjamin J. Ainscough, Lana M. Sheta, Shahil P. Pema, Alina D. Schmidt, Megan Richters, Kelsy C. Cotto, Arpad M. Danos, Cody Ramirez, Zachary L. Skidmore, Nicholas C. Spies, Jasreet Hun
Genetics in Medicine.2019; 21(4): 972. CrossRef - A DNA pool of FLT3-ITD positive DNA samples can be used efficiently for analytical evaluation of NGS-based FLT3-ITD quantitation - Testing several different ITD sequences and rates, simultaneously
Zoltán A. Mezei, Dávid Tornai, Róza Földesi, László Madar, Andrea Sümegi, Mária Papp, Péter Antal-Szalmás
Journal of Biotechnology.2019; 303: 25. CrossRef - Pharmacogenomic Testing: Clinical Evidence and Implementation Challenges
Catriona Hippman, Corey Nislow
Journal of Personalized Medicine.2019; 9(3): 40. CrossRef - Cancer Panel Assay for Precision Oncology Clinic: Results from a 1-Year Study
Dohee Kwon, Binnari Kim, Hyeong Chan Shin, Eun Ji Kim, Sang Yun Ha, Kee-Taek Jang, Seung Tae Kim, Jeeyun Lee, Won Ki Kang, Joon Oh Park, Kyoung-Mee Kim
Translational Oncology.2019; 12(11): 1488. CrossRef - Analytical Evaluation of an NGS Testing Method for Routine Molecular Diagnostics on Melanoma Formalin-Fixed, Paraffin-Embedded Tumor-Derived DNA
Irene Mancini, Lisa Simi, Francesca Salvianti, Francesca Castiglione, Gemma Sonnati, Pamela Pinzani
Diagnostics.2019; 9(3): 117. CrossRef - Benchmark Database for Process Optimization and Quality Control of Clinical Cancer Panel Sequencing
Donghyeong Seong, Jongsuk Chung, Ki-Wook Lee, Sook-Young Kim, Byung-Suk Kim, Jung-Keun Song, Sungwon Jung, Taeseob Lee, Donghyun Park, Byoung-Kee Yi, Woong-Yang Park, Dae-Soon Son
Biotechnology and Bioprocess Engineering.2019; 24(5): 793. CrossRef - Use of the Ion PGM and the GeneReader NGS Systems in Daily Routine Practice for Advanced Lung Adenocarcinoma Patients: A Practical Point of View Reporting a Comparative Study and Assessment of 90 Patients
Simon Heeke, Véronique Hofman, Elodie Long-Mira, Virginie Lespinet, Salomé Lalvée, Olivier Bordone, Camille Ribeyre, Virginie Tanga, Jonathan Benzaquen, Sylvie Leroy, Charlotte Cohen, Jérôme Mouroux, Charles Marquette, Marius Ilié, Paul Hofman
Cancers.2018; 10(4): 88. CrossRef - Use of the Ion AmpliSeq Cancer Hotspot Panel in clinical molecular pathology laboratories for analysis of solid tumours: With emphasis on validation with relevant single molecular pathology tests and the Oncomine Focus Assay
Ahwon Lee, Sung-Hak Lee, Chan Kwon Jung, Gyungsin Park, Kyo Young Lee, Hyun Joo Choi, Ki Ouk Min, Tae Jung Kim, Eun Jung Lee, Youn Soo Lee
Pathology - Research and Practice.2018; 214(5): 713. CrossRef - Recent Advancement of the Molecular Diagnosis in Pediatric Brain Tumor
Jeong-Mo Bae, Jae-Kyung Won, Sung-Hye Park
Journal of Korean Neurosurgical Society.2018; 61(3): 376. CrossRef - The long tail of molecular alterations in non-small cell lung cancer: a single-institution experience of next-generation sequencing in clinical molecular diagnostics
Caterina Fumagalli, Davide Vacirca, Alessandra Rappa, Antonio Passaro, Juliana Guarize, Paola Rafaniello Raviele, Filippo de Marinis, Lorenzo Spaggiari, Chiara Casadio, Giuseppe Viale, Massimo Barberis, Elena Guerini-Rocco
Journal of Clinical Pathology.2018; 71(9): 767. CrossRef - Clinical laboratory utilization management and improved healthcare performance
Christopher Naugler, Deirdre L. Church
Critical Reviews in Clinical Laboratory Sciences.2018; 55(8): 535. CrossRef - Development of HLA-A, -B and -DR Typing Method Using Next-Generation Sequencing
Dong Hee Seo, Jeong Min Lee, Mi Ok Park, Hyun Ju Lee, Seo Yoon Moon, Mijin Oh, So Young Kim, Sang-Heon Lee, Ki-Eun Hyeong, Hae-Jin Hu, Dae-Yeon Cho
The Korean Journal of Blood Transfusion.2018; 29(3): 310. CrossRef - Value-based genomics
Jun Gong, Kathy Pan, Marwan Fakih, Sumanta Pal, Ravi Salgia
Oncotarget.2018; 9(21): 15792. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
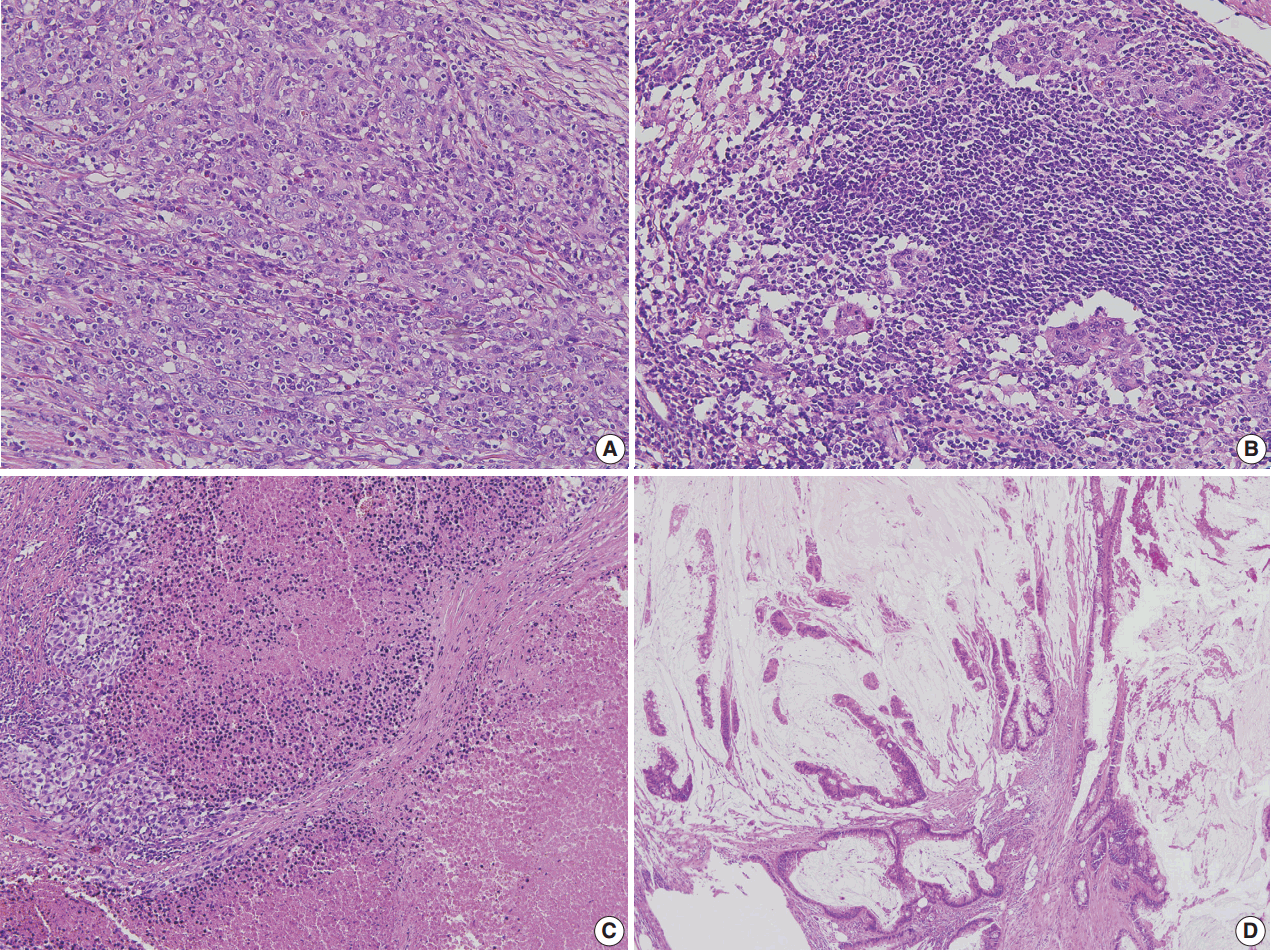
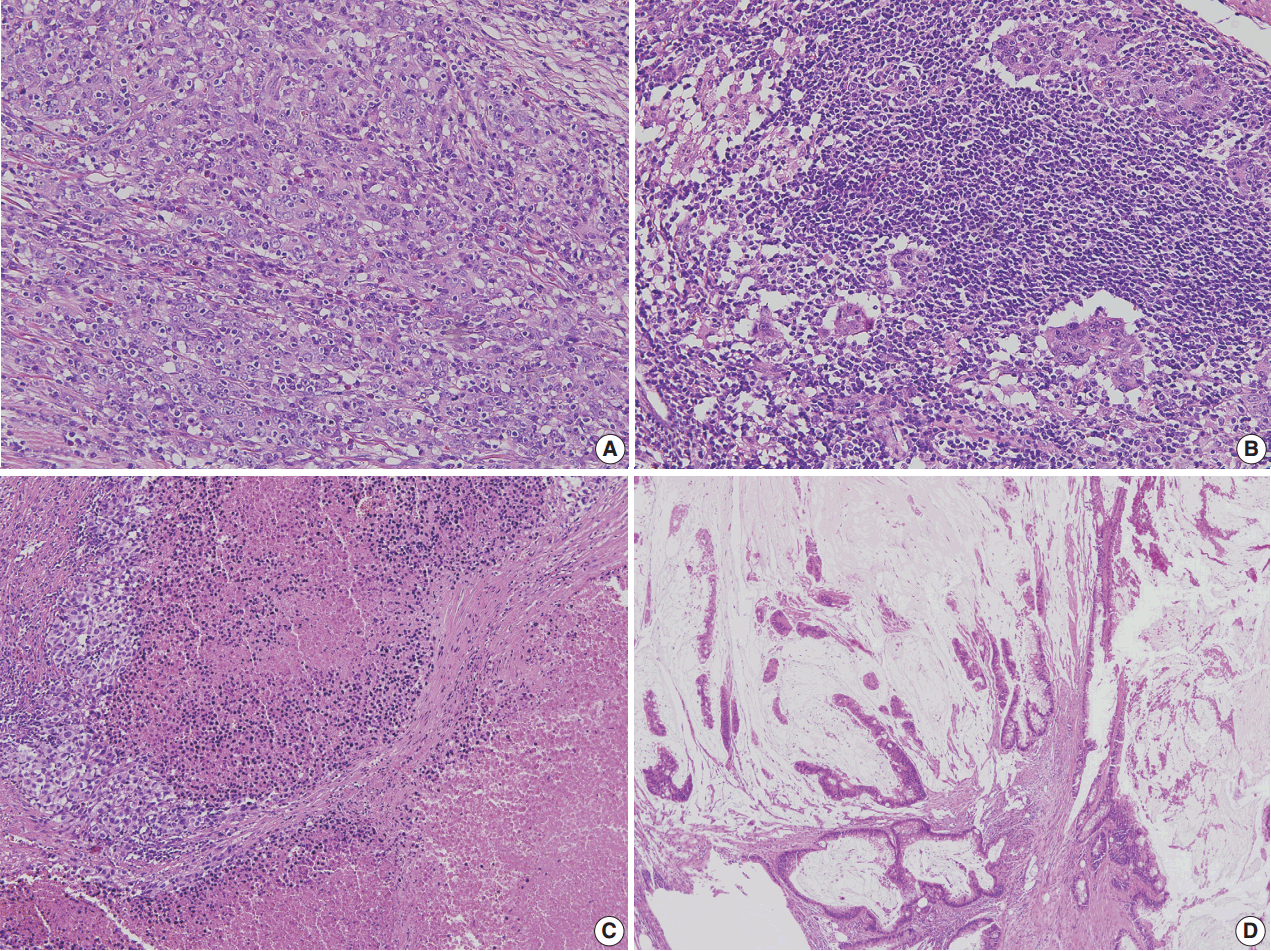
- Figure

Fig. 1.
| Subject | Database/Algorithm | URL |
|---|---|---|
| Germline polymorphism | dbSNP, dbVar | |
| 1000 Genomes Project | ||
| ExAC | ||
| Cancer-specific somatic variants | COSMIC | |
| cBioPortal | ||
| My Cancer Genome | ||
| CIViC | ||
| Personalized Cancer Therapy, MD Anderson Cancer Center | ||
| Genotype-phenotype association, not limited to cancer | ClinVar | |
| Human Gene Mutation Database | ||
| Leiden Open Variation Database | ||
| In silico functional prediction | dbNSFP (pre-computed in silico functional prediction and annotation of non-synonymous SNVs) | |
| Ensembl Variant Effector Predictor |
| Element | Example |
|---|---|
| Patient identification | Registration number, age, gender, ordering physician |
| Specimen type | FFPE, fresh frozen |
| Pathologic diagnosis | Lung adenocarcinoma, clorectal cancer |
| Tissue sample identification | Specimen number, block number |
| Important dates | Date on reception or on report |
| Percentage of tumor nuclei of the sample used | 30% |
| Variants found | Variant details according to HGVS mutation nomenclature |
| Version of reference genome used | hg19 build 36 |
| NGS method used | Amplicon-based or hybridization capture-based |
| Key quality control metrics | Mean target coverage, percentage of selected bases, duplication rate |
| Genes or genomic regions included in the panel | Exonic regions of gene A, B, C, etc. |
| Interpretation and summary | EGFR L858R variants predict response to EGFR tyrosine kinase inhibitors (Erlotinib or Gefitinib) |
| Type of material | Advantage | Disadvantage |
|---|---|---|
| Genomic DNA from cell line | Large amount available | May have heterogeneity associated with cell line maintenance |
| Similar complexity to patient’s DNA | Possible genomic instability over time | |
| May have well-characterized variants | ||
| Genomic DNA from patient’s sample | Identical condition to real samples | Not necessarily renewable |
| Limited amount of DNA | ||
| Well characterized genetic information may be limited | ||
| Synthetic DNA | Can synthesize a broad range of sequences and variations | Does not represent actual human cancer genome |
| May not perform as actual human cancer DNA due to differences in sequence complexity | ||
| Can make sequence templates with complex regions (deletions or duplications) | ||
| Will not cover all regions of tested genome | ||
| Large amount available | May exhibit higher variant calls due to errors in synthesis | |
| Electronic reference data files | Can engineer any wanted sequence files | Reference only for data analysis step |
| Requires many reference datasets to mimick many types of sequence data | ||
| Data files may not be interoperable among different platforms |
| Item | Checklist | Consequences of non-conformity | Improvement suggestions |
|---|---|---|---|
| Tissue sample adequacy | Criteria for inadequate specimen | Testing inadequate specimens may lead to a waste of time and money or depletion of available samples. | Check sample adequacy rigorously before testing. |
| Minimum tumor content | Request further sampling in case of inadequate samples. | ||
| Appropriate sample handling including fixation and transportation | Inadequate amount or tumor content can lead to false-negative test results. | ||
| Nucleic acid extraction | DNA quantity and quality in terms of amplifiable DNA | DNA with suboptimal quality may inhibit sequencing reaction. | Failed samples should be reported as such and further material might be requested with specified requirement. |
| Small amount or fragmentation of DNA may lead to poor quality sequencing data with insufficient or uneven coverage and/or high duplication rate. | |||
| Trying another validated extraction method may often helps. | |||
| Sample identification | Sample identity tracking throughout all steps | Misidentification of samples could lead to incorrect patient management. | If there is any concern about sample identity, starting over from DNA extraction may be necessary. |
| Introduction of polymorphic SNP markers into gene panel and running another genotyping method with the same marker set might be helpful. | |||
| Library preparation | Minimum library concentration | Poor sequencing library may lead to insufficient or uneven coverage. | Consider modification of library preparation method or an alternative method to verify any uncertain results. |
| Libraries with poor complexity or bias may result in false-negatives. False-positives may also occur due to potential amplification bias. | |||
| Sequencing | Criteria for minimum sequencing depth and other quality metrics (% reads mapped to target regions, % of targets with specified coverage, duplication rate) | Inadequate coverage is associated with higher levels of uncertainty of the test results. | Repeat sequencing with existing library or start over from DNA extraction step. |
| Genomic regions with insufficient local coverage may lead to inaccurate results for variants located in those regions. | Verification of uncertain results with another method may be helpful, especially, in case of actionable variants. | ||
| Variant detection and review | Variant allele frequency, local sequencing depth and quality score | Failure to filter out sequencing artifacts may lead to false-positive results. | Manually review of clinically important variants even if computational algorithms called no mutation on them. |
| Presence of the same variant in forward and reverse strands | Clinically important variants may sometimes be missed. | Any ambiguous or unexpected results should be reviewed by laboratory scientists and pathologists. | |
| Mapping quality of sequencing reads | Verify variants with another method, if applicable. | ||
| Potential sequencing artifacts | |||
| Bioinformatics | Correct pipeline and version | Using outdated or inadequate software can lead to false-positive or false-negative results. | Update software on a regular basis. |
| Appropriate version and build of human reference sequence | |||
| Cross-contamination? | |||
| Reporting | Endorsed by an authorized competent pathologist? | Variants with clinical significance may be reported erroneously, leading to inappropriate treatment. | Responsible pathologists should be given enough time and opportunities for education and training. |
| Quality metrics | General considerations upon validation | Actions to be considered during quality controls |
|---|---|---|
| Depth of coverage | Minimum coverage threshold required for desired sensitivity and specificity across targeted regions should be established. | When the coverage threshold could not be met in a suspicious variant call, additional validation by an alternate method (e.g., |
| Sanger sequencing) should be considered. | ||
| Uniformity of coverage | Coverage across targeted regions should be as uniform as possible to produce reliable results across all targeted genomic regions. | Uniformity of coverage often decreases with samples having low DNA amount or highly degraded DNA. |
| When coverage uniformity significantly deviates from that established during initial validation, this may indicate potential analytical errors. | ||
| GC bias | GC content affects the efficiency of the sequencing reactions and will affect the uniformity of coverage. | GC bias should be monitored in every run to detect any change in test performance. |
| The amount of GC bias should be established across the targeted genomic regions. | ||
| Base call quality scores | Informatics filters should be in place to eliminate any reads having a base call quality score below the acceptable quality score. | Quality scores are not readily comparable from one sequencing platform to another. |
| Mapping quality | During validation, it is important to make sure that the test analyzes the reads that map specifically to the targeted genomic regions. | The proportion of reads that do not map to targeted regions should be monitored during each run. |
| Informatics filters should be in place to eliminate any reads that map to non-targeted regions. | Poor mapping quality may come from non-specific amplification, capture of off-target DNA, or contamination. | |
| Proportion of duplicated reads | Informatics filters should be in place to eliminate duplicate reads resulting from clonal amplification during alignment. | The amount of duplicate reads should be monitored to prevent skewing of allelic fractions. |
This list is not comprehensive and only provides some examples. All websites were last accessed on January 3, 2017. ExAC, Exome Aggregation Consortium; COSMIC, Catalog of Somatic Mutations in Cancer; CIViC, Clinical Interpretations of Variants in Cancer; NSFP, nonsynonymous functional prediction; SNV, single nucleotide variation.
NGS, next-generation sequencing; FFPE, formalin-fixed paraffin embedded; HGVS, Human Genome Variation Society; EGFR, epidermal growth factor receptor.
Adapted by permission from Macmillan Publishers Ltd: [Nature Biotechnology] Gargis NGS, next-generation sequencing.
NGS, next-generation sequencing; QC, quality control.
Adapted by permission from Macmillan Publishers Ltd: [Nature Biotechnology] Gargis NGS, next-generation sequencing.

 E-submission
E-submission