Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 53(1); 2019 > Article
-
Review
Artificial Intelligence in Pathology -
Hye Yoon Chang
 , Chan Kwon Jung1
, Chan Kwon Jung1 , Junwoo Isaac Woo
, Junwoo Isaac Woo , Sanghun Lee
, Sanghun Lee , Joonyoung Cho
, Joonyoung Cho , Sun Woo Kim
, Sun Woo Kim , Tae-Yeong Kwak
, Tae-Yeong Kwak
-
Journal of Pathology and Translational Medicine 2019;53(1):1-12.
DOI: https://doi.org/10.4132/jptm.2018.12.16
Published online: December 28, 2018
Deep Bio Inc., Seoul, Korea
1Department of Hospital Pathology, College of Medicine, The Catholic University of Korea, Seoul, Korea
- Corresponding Author Tae-Yeong Kwak, PhD Deep Bio Inc., 1201 Hanwha Bizmetro, 242 Digital-ro, Guro-gu, Seoul 08394, Korea Tel: +82-70-7703-4746 Fax: +82-2-2621-2223 E-mail: tykwak@deepbio.co.kr
© 2019 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
- As in other domains, artificial intelligence is becoming increasingly important in medicine. In particular, deep learning-based pattern recognition methods can advance the field of pathology by incorporating clinical, radiologic, and genomic data to accurately diagnose diseases and predict patient prognoses. In this review, we present an overview of artificial intelligence, the brief history of artificial intelligence in the medical domain, recent advances in artificial intelligence applied to pathology, and future prospects of pathology driven by artificial intelligence.
- Since the earliest stage of modern AI research, substantial efforts have been made in the medical domain. A script-based chatbot named ELIZA was proposed in 1966 [20]. ELIZA’s most famous script, DOCTOR, could interact with humans as a Rogerian psychotherapist. A biomedical expert system, MYCIN, presented in 1977, could analyze infectious symptoms to derive causal bacteria and drug treatment recommendations [21]. Later, in 1992, the probabilistic reasoning-equipped PATHFINDER expert system was developed for hematopathology diagnosis, to deal with uncertain biomedical knowledge efficiently [22,23].
- Before the era of DL, several ML methods have been used widely in the medical domain. Moreover, the invention of digital medical imaging such as digital X-ray imaging, computed tomography and magnetic resonance imaging enabled computerized image analysis, where AI achieved another success in the medical domain. In 1994, Vyborny and Giger [24] reviewed the efforts to use ML algorithms featuring computer vision in several mammography analysis tasks, including microcalcification detection, breast mass detection and differentiation of benign from malignant lesions. They demonstrated the efficacy of computer-aided detection (CAD) by comparing the performance of radiologists with CAD to that of radiologists only. Later, in 2001, Kononenko [25] overviewed the typical ML methods such as decision trees, Bayesian classifiers, neural networks, and k nearest neighbor (k-NN) search, then reviewed their use in medical diagnosis and proposed evaluation criteria including performance, transparency, explainability and data resiliency. In 2003, however, Baker et al. [26] pointed out that the performance of commercial CAD systems was still below the expectation (max case sensitivity 49%) in detecting architectural distortion of breast mammography.
- After the success of deep CNN in image classification, a wide range of attempts were made to apply DL to medicine. A notable success was the work of Gulshan et al. [27] in 2016, where retinal fundus images were analyzed by a CNN-based DL model to detect diabetic retinopathy lesions, achieving an area under the receiver operating characteristic curve (AUC) of 0.991, sensitivity of 97.5% and specificity of 93.4% in the high sensitivity setting, measured on the EyePACS-1 data set. In 2017, Litjens et al. [28] reviewed major DL methods suitable for medical image analysis and summarized more than 300 contributions in the neuro, retinal, pulmonary, breast, cardiac, abdominal, and musculoskeletal areas as well as in the digital pathology domain; contributions were well categorized according to their inherent type of image analysis: classification, detection, segmentation, registration, etc. Kohli et al. [29] presented another review on the application of ML to radiology research and practice, where transfer learning and data augmentation were emphasized as a viable solution to datalimited situations. Shaikhina and Khovanova [30] proposed another solution for a similar situation; their proposed solution incorporates multiple runs and the surrogate data test, which exploits statistical tools to guide the trained ML model having better model parameters and not being overfitted to a small training data set.
- Genomics and molecular biology have been strongly connected to the medical domain since genome sequencing became real. Next-generation sequencing (NGS) technology allows a whole genome sequence to be translated into text composed of ATCG, so that necessary computational analysis can be done for disease diagnosis and therapeutic decision making. In 2016, Angermueller et al. [31] reviewed DL methods and their application to genomic and biological problems such as molecular trait prediction, mutation effect prediction, and cellular image analysis. They thoroughly reviewed the whole process used to apply DL to their problems, from data acquisition and preparation to overfit avoidance and hyperparameter optimization. Torkamani et al. [32] presented a review of high-definition medicine, which is applied to personalized healthcare by using several kinds of big data, including DNA sequences, physiological and environmental monitoring data, behavioral tracking data and advanced imaging data. Surely, DL techniques can help in analyzing those big data datasets in parallel, to provide exact diagnosis and personalized treatment. Another review was done in 2018 by Wainberg et al. [33] on the use of DL in various biomedical domains, including quantitative structureactivity relationship modeling for drug discovery and identification of pathogenic variants in genome sequences. They re-emphasized the importance of the performance, transparency, model interpretability and explainability of DL methods, in earning the trust of stakeholders gaining adoption. Besides these reviews, there exist two notable contributions for genetic variants. Xiong et al. [34] presented a computational model for gene splicing, which can predict the ratio of transcripts with the central exon spliced in, within the whole set of transcripts spliced from any given sequence containing an exon triplet. Recently an award-winning deep CNN-based variant caller named DeepVariant was announced [35], which is able to call genetic variation in aligned NGS read data by learning on images created upon the read pileups around putative variant sites.
- Another type of medical data to be analyzed is electronic health records (EHR). Rajkomar et al. [36] recently published their work building a DL model that predicts multiple medical events, including in-hospital mortality, unplanned readmission, and prolonged length of stay, entirely from raw EHR records based on the Fast Healthcare Interoperability Resources format. Their model could accurately predict mortality events, with an AUC of 0.90 at patients’ admission, and even with an AUC of 0.87 at 24 hours before admission to the hospital. EHR data can be used in the prediction of other types of events, e.g., outcome of a patient biopsy, which could be predicted with AUC 0.69 in the work of Fernandes et al. [37]
- Besides the analytical diagnostic tasks, AI has been tried in other areas, for example, an intelligent assistant named Secretary-Mimicking Artificial Intelligence that helps in the execution of a pathology workflow was presented by Ye [38]. Treatment decision is another important factor in patient healthcare, from both prognostic and financial perspectives. Markov decision analysis is an effective tool in such situations, which was used to solve the cardiological decision problem in the work presented by Beck et al. [39] Schaefer et al. [40] reviewed the medical treatment modeling using the Markov decision process, which is a modeling tool that fits well in the optimization of sequential decision making and is strongly related to reinforcement learning [41].
HISTORY OF ARTIFICIAL INTELLIGENCE IN MEDICINE
- Microscopic morphology remains the gold standard in diagnostic pathology, but the main limitation to morphologic diagnosis is diagnostic variability in bearing error among pathologists. The Gleason grading system is one of the most important prognostic factors in prostate cancer. However, significant interobserver variability has been reported when pathologists have used the Gleason grading system [42,43]. In order to get a consistent and possibly more accurate diagnosis, it is natural to introduce algorithmic intelligence in the pathology domain, at least in the morphological analysis of tissues and cells. With the help of digital pathology equipment varying from microscopic cameras to whole slide imaging scanners, morphology-based automated pathologic diagnosis has become a reality. In this review, we focus on morphology-based pathology: diagnosis and prognosis based on the qualitative and quantitative assessment of pathology images. Typical digital image analysis tasks in diagnostic pathology involve segmentation, detection, and classification, as well as quantification and grading [44]. We briefly introduce typical techniques used for AI in digital pathology and a few notable research studies per disease. The list of studies reviewed in this paper is given in Table 2.
ARTIFICIAL INTELLIGENCE APPLICATION IN PATHOLOGY
- Digital pathology images used in AI are mostly scanned from H&E stained slides. Pathology specimens undergo multiple processes, including formalin fixation, grossing, paraffin embedding, tissue sectioning, and staining. Each step of the process and the different devices and software used with the digital imaging scanners can affect aspects of the quality of the digital images, such as color, brightness, contrast, and scale. For the best results, it is strongly recommended to alleviate the effect of these variations before using the images in automated analysis work [45]. Normalization is one of the techniques used to reduce such variations. Simple linear range normalization based on the equation [vnew = (vold-a)/fscale + b] is generally used for the pixel values in grayscale images, or for each channel of color images [47,60]. Scale normalization has not been reported in related works, as they all have used a single image acquisition device, e.g., a certain microscopic camera or digital slide scanner. When multiple image acquisition devices are used, scale normalization is of concern, since images acquired from different devices can have different pixel sizes, even at the same magnification level.
- Detecting the region-of-interest (ROI) has been done by combining several computer vision operations, such as color space conversion, image blurring, sharpening, edge detection, morphological transformation, pixel value quantization, clustering, and thresholding [67]. Color space conversion is often done before pixel clustering or quantization, to separate chromatic information and intensity information [53]. Another type of color space conversion targets direct separation of color channels for hematoxylin (H), eosin (E) and diaminobenzidine from stained tissue images to effectively obtain nuclei area [57,59,66,68]. Thresholding based on a certain fixed value leads to low-quality results when there are variations in luminance in the input images. Adaptive thresholding methods like hysteresis thresholding and Otsu’s method can generate better thresholding results [47,53,59,69]. Recently, pixel-wise or patch-wise classifiers based on CNN have been used widely in ROI detection [44,49-51,54-56,58,65], where a deep CNN is trained to classify the type of target pixel or patch centered on the larger input image patch in a sliding window manner. Semantic segmentation CNN is another recent trend for this task [65,70,71], which can detect multiple ROIs in a given image without sliding window operation, resulting in much faster speed.
- In the development of a CNN-based automated image analysis, data-limited situations are common in the medical domain, because it is very costly and time-consuming to build a large amount of annotated, high-quality data [45]. As previously mentioned, transfer learning and data augmentation should be incorporated to get a better result. In transfer learning, convolutional layer parameters of a CNN, pre-trained with a well-known dataset like ImageNet, are imported into the target CNN as layer initialization, while later layers like fully connected layers or deconvolutional layers are initialized randomly [62,70,71]. Additional training steps can update all of the layer parameters, including the imported ones, or only the parameters of the layers that were randomly initialized. With sufficient data, building a model without transfer learning is reported to give better performance [54].
- A common strategy of image data augmentation is, for the given image, applying various transformations that do not alter the essential characteristics; such transformations include rotation (90°, 180°, and 270°), flipping (horizontal/vertical), resizing, random amounts of translation, blurring, sharpening, adding jitters in color and/or luminance, contrasting histogram equalization, etc [47,51,52,56,60-63]. Another type of augmentation relates to the patch generation strategy; applying large medical images directly to the CNN is impractical. From a large pathological image, with a size between 1024 × 1024 (camera) and > 104 × 104 (scanner) pixels, smaller patches with sizes between 32 × 32 and 512 × 512 pixels are retrieved for use in training and inference of CNNs. Instead of using the pre-generated set of image patches through the whole training process, resampling patches during each training epoch can introduce more variance in training data to reduce the chance of overfitting [60].
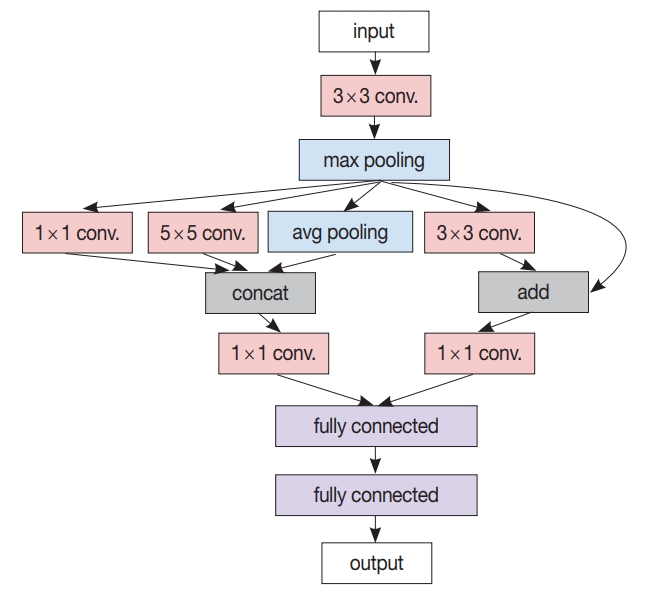
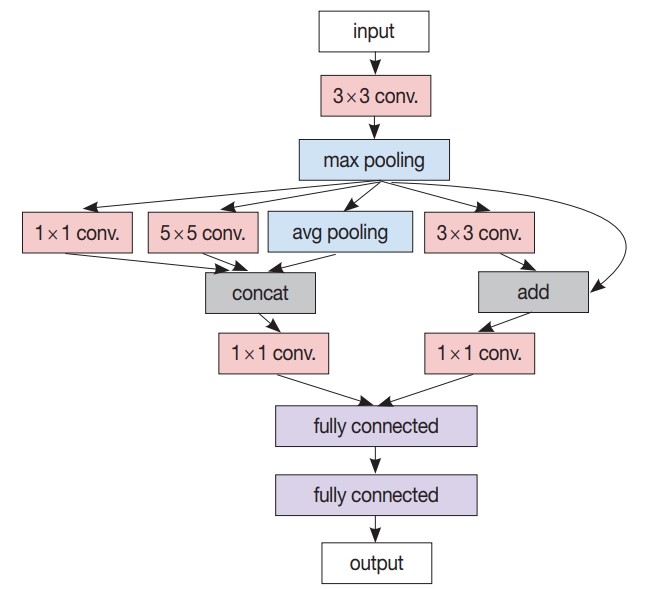
- After the patch-level CNN is trained, another ML model is often developed for the whole image level decision. In this case, a patch-level decision is made for every single patch in the training images to generate heatmap-like output, from which several features are extracted via conventional image analysis methods. Then, collected feature values for the training images are fed into the target image level ML model. An example workflow for developing and using this two-stage pathology AI is depicted in Fig. 3.
TYPICAL TECHNIQUES
- CNN-based breast cancer diagnosis was tried with fine needle aspiration (FNA) cytology images [46], optical coherence tomography (OCT) images [48], and H&E stained tissue images [49], each with varying numbers of data points and model structures. A total of 175 cytology images captured by a microscopic camera at 40 × magnification level were manually split into 918 ROIs, 256 × 256 pixels in size, where each ROI had multiple cells [46]. A CNN was trained to determine the malignancy of a given ROI, and the cytological image was classified as malignant when > 30% of the ROIs in the image were malignant. The reported accuracy was 89.7%, which was far inferior to the 99.4% accuracy of a random forest classifier with 14 hand-crafted features. In order to attempt an automated intraoperative margin assessment, 4,921 frame images from the frozen section OCT were used, from which patches 64 × 64 pixels in size were extracted, resized to 32 × 32 pixels, and used for training and evaluation [48]. Patch-level CNN performance was measured, giving an accuracy of 95.0% and AUC of 0.984 in the best setting. In another study, 2,387 H&E stained breast biopsies were scanned at a magnification of 20 × [49]. Multiple CNNs were used in this study: the first CNN classified each image pixel as fat, stroma, or epithelium; the second CNN predicted whether each stromal pixel was associated with a cancer; and the third CNN determined the whole-slide-level malignancy. The reported slide level AUC was 0.962. A notable result is that, while the CNNs were trained with stromal tissues in benign slides and invasive cancer slides only, the predicted cancer association probability of the stroma near the ductal carcinoma in situ (DCIS) lesion properly related to the severity of DCIS. CNN-based lymph node metastasis detection was also tried with a different model and dataset [47,50]. Conditional random field was adopted on top of convolutional layers in order to regulate the metastasis prediction [47]. From the whole slide images (WSIs) in the CAMELYON16 dataset [72], benign and tumor image patches 768 × 768 pixels in size were sampled to train and validate the model, giving patch-level accuracy of 93.8% after incorporating data augmentation methods. In another study, 271 WSIs scanned at a magnification of 20 × were used in developing a CNN-based model for detecting micro- or macro-metastasis-free slides [50]. Region-level annotations on training images were utilized. Slide-level metastasis detection was performed after metastasis probability map generation by patch-level CNN, incorporating probability thresholding (> 0.3) and connected component analysis to remove small lesions (< 0.02 mm diameter), resulting in a detection AUC of 0.90. Mitosis detection was tried with a CNN that decides whether the center of the given image is mitotic or not [51], trained and evaluated with 50 images from five biopsy slides containing about 300 mitoses total, adopting data augmentation techniques including patch rotation and flipping. In the evaluation, a mitosis probability map was created for the given image, and pixels with locally maximal probabilities were considered as mitotic, resulting in detection F1-score 0.782.
- Automatic lung cancer subtype determination was tried with FNA cytology images and H&E stained WSIs [52,54]. A total of 298 images from 76 cases acquired using a microscopic camera at 40 × magnification level were utilized in developing a CNN receiving 256 × 256 pixel images as input; the dataset comprised 82 adenocarcinomas, 125 squamous cell carcinomas, and 91 small cell carcinomas [52]. Data augmentation techniques like rotation, flipping, and Gaussian filtering were adopted to enhance the classification accuracy from 62.1% to 71.1%. A total of 1,635 WSIs from the The Cancer Genome Atlas (TCGA) [73] dataset were used in detection of lung cancer type with CNN [54]. Each input patch (512 × 512 pixels) was classified as adenocarcinoma, squamous cell carcinoma or benign, and then the averaged probability of non-benign patches was used in the slide-level decision, resulting in slide level classification AUC of 0.97, which is much superior to the previous SVM-based approach [53]. Moreover, by using the multi-task transfer learning approach, mutations of six genes including KRAS, EGFR, and STK11 were independently able to be determined on the input WSI of lung adenocarcinoma patches. The mutation detection had an AUC of 0.86 for STK11 and an AUC of 0.83 for EGFR.
- Prostate cancer diagnosis has been one of the most active fields in adopting DL because of its large dependence on tissue morphology. Prostatic tissues from various sources have been used in malignancy and severity decisions [50,55-58]. In one study, 225 prostate needle biopsy slides were scanned at 40 × magnification, and malignant regions were annotated in developing a cancer detector [50]. A CNN-based patch-level cancer detection was performed for every overlapping patch in a slide to generate a probability map, and a cumulative probability histogram was created and analyzed in slide-level malignancy determination (AUC 0.99). In another study, 12,160 needle biopsy images were utilized in developing a CNN-based slide-level malignancy detector [55]. To train a patch-classifying CNN with no patch/regionlevel manual annotation, multiple instance learning was used; with a large number of WSIs (> 8,000), the result was useful (AUC 0.98). A total of 886 tissue microarray (TMA) samples were used in a trial of automated Gleason scoring [56], where 508 TMA images for training were manually segmented into combinations of benign, Gleason pattern 3, 4, and 5; 133 TMA images were used for tuning and 245 images were used for validation. The TMA level score was determined by the two most dominant patterns measured from the per-pattern probability maps generated by a trained patch-level CNN classifier. In grading the validation set, Cohen’s kappa between two pathologists was 0.71, while those between the model and each of the two pathologists were 0.75 and 0.71. 342 cases from TCGA, teaching hospital and medical lab were utilized in training automated Gleason scoring system [58], where CNN and k-NN classifier were ensembled. A total of 912 slide images were annotated with the region level to be used in training CNN to generate a pattern map for a given slide image; 1,159 slides were used to train the k-NN classifier that determines the Gleason group for the given pattern map statistics. The reported grading accuracy measured on 331 slides was 0.70, while the average accuracy of 29 general pathologists was 0.61, which is superior to the previous TCGA-based result that showed 75% accuracy in discriminating Gleason score 3 + 4 and 4 + 3 [57].
- An automated determination of brain cancer severity was tried with TCGA brain cancer data [59]. A cascade of CNNs was used: an initial CNN trained with 22 WSIs for discriminating between glioblastoma (GBM) and low-grade glioma (LGG), and a secondary CNN trained with an additional 22 WSIs for discriminating between LGG grades 2 and 3. Each H&E-stained RGB color image was transformed into an H-stained channel and an E-stained channel, and only the H-stained channel was used for further analysis. The first CNN showed GBM/LGG discrimination accuracy of 96%, but the LGG grade discrimination was not so successful (71%). Survival analysis using CNN was also tried [60]. Again, 1,061 WSIs from TCGA dataset were used. For each training epoch, 256 × 256 pixel patches were sampled from manually identified, 1,024 × 1,024 pixel ROIs. At diagnosis, ROI-wise risk was determined as the median risk of nine patches sampled from the ROI, and the sample-level risk was determined as the second highest risk among ROI risks. The measured c-index of this kind of survival analysis was 0.75, which was elevated to 0.80 by modifying the CNN to receive the mutation information at its fully connected layer.
- Ovarian cancer subtype classification based on CNN was tried [61]. 7,392 images were generated by splitting and cropping the original images acquired by the microscopic camera at 40 × magnification level. Rotation and image quality enhancement were used in the data augmentation phase, which enhanced the classification accuracy from 72.8% to 78.2%. Cervical cancer diagnosis on cytological images was also tried [62]. Without cell-wise segmentation, nuclei-centered cell patches were sampled from the original cytology image, followed by augmentation operations like rotation and translation. Convolutional layer parameters that were trained by using ImageNet data were transferred to actual CNN. Herlev and HEMLBC datasets were used in evaluation, giving 98.3% and 98.6% accuracy, respectively, in five-fold cross-validation. Red blood cell (RBC) classification is crucial in sickle cell disease diagnosis. A CNN-based automatic RBC classification was tried [63], where 7,206 cell patches were generated from 434 microscopic images and used for training and testing of the classifier. Rotation and flipping were used to augment training data. Five-fold cross-validation showed an average accuracy of 89.3% in five-class coarse classification, and 87.5% in eight-class refined classification. A total of 469 TMA cores from the gastric cancer patients were used in a CNN-based survival analysis [64]. CD8 and Ki67 immunostained images were acquired and fed into separate patch-wise risk-predicting CNNs for each stain. From the differential analysis between the low-risk group and the high-risk group, it was claimed that the density of CD8 cells was largely related to the risk level.
EXAMPLES OF PATHOLOGY ARTIFICIAL INTELLIGENCE
- We have provided an overview of various medical applications of AI technology, especially in pathology. It is encouraging that the accuracy of automated morphological analyses has improved due to DL technology. The pathologic field in AI is expanding to disease severity assessment and prognosis prediction. Although most AI research in pathology is still focused on cancer detection and the grading of tumors, pathological diagnosis is not simply a morphological diagnosis, but is a complex process of evaluation and judgment of various types of clinical data that deal with various organs and diseases. A large amount of data, including genetic data, clinical data, and digital images, is needed to develop AI that covers the range of clinical situations. There are a number of public medical databases, including TCGA, and a number of studies have been done based on those databases. They provide a good starting point in researching and developing a medical AI, but it requires much more high-quality data; e.g., detailed annotations on a large number of pathology images, created and validated by several experienced pathologists, are necessary to develop a pathology-image-analyzing AI that is comparable to human pathologists.
- There are difficulties in constructing such high-quality data in reality, largely due to the protection of privacy, proprietary techniques, and the lack of funding and pathologists to participate in the annotation process. To overcome this data insufficiency, as we have mentioned earlier, several techniques have been introduced, such as transfer learning and data augmentation. Still, these techniques are sub-optimal; transfer learning cannot guarantee the optimal convolutional filters specific for the task, and data augmentation cannot deal with the unseen data and patterns. The ultimate solution is to construct a large amount of thoroughly labeled and annotated medical data, through the cooperation of multiple hospitals and medical laboratories. To accelerate the construction of such a dataset, efficient tools for labeling and annotating are required, which can be assisted by another type of AI [45].
- Eventually, there will be a medical AI of the prognostic prediction model, combining clinical data, genetic data, and morphology. Also, a new grading system applicable to several tumors can be created by an AI model that has learned from the patient’s prognosis combined with a number of variables including morphology, treatment modality, and tumor markers, etc. This will also help to overcome the poor reproducibility and the variety of current grading and staging results among pathologists, leading to much better clinical outcomes for patients.
FUTURE PROSPECTS
Acknowledgments


| Author (year) | Disease | Data | Task | Model | Augmentation | Performance |
|---|---|---|---|---|---|---|
| Garud et al. (2017) [46] | Breast cancer | FNA cytology/175 (images) | Decision Benign/cancer | CNN | None | Image level decision acc. 89.7% |
| Li and Ping (2018) [47] | Lymph node metastasis | CAMELYON16/400 (WSIs) | Decision Yes/no | CNN + CRF | Color jitter, rotation, etc. | Patch level decision acc. 93.8% |
| Rannen Triki et al. (2018) [48] | Breast cancer | Frozen section OCT/4,921 (frames) | Decision Benign/cancer | CNN | None | Patch level decision acc. 94.96% |
| Ehteshami Bejnordi et al. (2018) [49] | Breast cancer | BREAST Stamp/2,387 (WSIs) | Decision Benign/cancer | CNN + CNN | None | WSI level decision AUC 0.962 |
| Litjens et al. (2016) [50] | Lymph node metastasis | Lymph node specimen/271 (samples) | Decision Yes/no | CNN | None | Sample level decision AUC 0.90 |
| Cires¸ an et al. (2013) [51] | Breast cancer | MITOS/300 mitosis in 50 images | Mitosis detection | CNN | Rotation, flip, etc. | Detection F1-score 0.782 |
| Teramoto et al. (2017) [52] | Lung cancer | FNA cytology/298 (images) | Classification | CNN | Rotation, flip, etc. | Overall classification acc. 71.1% |
| Adeno-Squamous cell | ||||||
| Small cell | ||||||
| Yu et al. (2016) [53] | Lung cancer | TCGA-LUAD/1,074 | Decision Benign/cancer | SVM | None | Patch level decision AUC 0.85 |
| TCGA-LUSC/1,111 | Survival analysis | |||||
| Stanford TMA/294 (samples) | ||||||
| Coudray et al. (2018) [54] | Lung cancer | TCGA lung cancer/1,635 (samples) | Classification | CNN | None | Overall classification AUC 0.97 |
| Adeno-Squamous cell | STK11 mutation decision AUC 0.85 | |||||
| Benign | ||||||
| Multi-task decision | ||||||
| Gene mutation | ||||||
| Campanella et al. (2018) [55] | Prostate cancer | Needle biopsy/12,160 (samples) | Decision Benign/cancer | CNN (MIL) | None | Sample level decision AUC 0.979 |
| Arvaniti et al. (2018) [56] | Prostate cancer | TMA/886 (samples) | Classification Gleason score | CNN +scoring rule | Rotation, flip, color jitter | Model-pathologist Cohen’s kappa 0.71 |
| Zhou et al. (2017) [57] | Prostate cancer | TCGA-PRAD/368 (cases) | Decision 3 + 4/4 + 3 | CNN | None | Sample level decision acc. 75% |
| Nagpal et al. (2018) [58] | Prostate cancer | TCGA-PRAD + others/train 1,226, test 331 (slides) | Classification Gleason group | CNN + k-NN | None | Overall classification acc. 70% |
| Survival analysis | C-index 0.697 | |||||
| Litjens et al. (2016) [50] | Prostate cancer | Needle biopsy / 225 (WSIs) | Decision Benign/cancer | CNN | None | Slide level decision AUC 0.99 |
| Ertosun and Rubin (2015) [59] | Brain cancer | TCGA-GBM & LGG (unknown size) | Classification | CNN + CNN | Color transform to H&E | GBM/LGG decision acc. 96% |
| GBM | LGG grade decision acc. 71% | |||||
| LGG grade 2 | ||||||
| LGG grade 3 | ||||||
| Mobadersany et al. (2018) [60] | Brain cancer | TCGA-GBM & LGG/1,061 (samples) | Survival analysis | CNN | Rotation, normalization | C-index 0.754 |
| Wu et al. (2018) [61] | Ovarian cancer | Biopsy/7,392 (images) | Classification Subtypes | CNN | Rotation, image enhancement | Overall classification acc. 78.2% |
| Zhang et al. (2017) [62] | Cervix cancer | HEMLBC/1,978 Herlev/917 (images) | Decision Benign/cancer | CNN | Rotation, translation, | Image level decision AUC 0.99 |
| Xu et al. (2017) [63] | Sickle cell disease | Red-blood cell/7,206 (patches) | Classification Cell types | CNN | Rotation, flip, translation, etc. | Cell level classification acc. 87.5% |
| Meier et al. (2018) [64] | Gastric cancer | TMA/469 (samples) CD8/Ki67 IHC | Survival analysis | CNN | None | Stratification by risk successful (p < .01) |
| Xie et al. (2016) [65] | - | Synthetic fluorescence microscopy cell/200 (images) | Cell counting | CNN | None | Mean absolute error < 2% |
| Tuominen et al. (2010) [66] | - | IHC stained breast cancer slides/100 | Cell counting | Comp. vision | None | Correlation coefficient 0.98 |
CNN, convolutional neural network; MIL, multiple instance learning; SVM, support vector machine; AUC, area under receiver operating characteristic curve; k-NN, k-nearest neighbor; WSI, whole slide image; CRF, Conditional random field; TCGA, The Cancer Genome Atlas; TMA, tissue microarray; IHC, immunohistochemistry; GBM, glioblastoma multiforme; LGG, lower grade glioma.
- 1. McCorduck P. Machines who think: a personal inquiry into the history and prospects of artificial intelligence. Natick: A.K. Peters, 2004.
- 2. Turing AM. I. Computing machinery and intelligence. Mind 1950; 59: 433-60. ArticlePDF
- 3. Searle JR. Minds, brains, and programs. Behav Brain Sci 1980; 3: 417-24. Article
- 4. Russell SJ, Norvig P. Artificial intelligence: a modern approach. Upper Saddle River: Prentice Hall, 2003.
- 5. Artificial intelligence [Internet] Wikipedia, 2018 [cited 2018 Dec 9]. Available from: https://en.wikipedia.org/wiki/Artificial_intelligence.
- 6. Mortensen TL, Watt DL, Leistritz FL. Loan default prediction using logistic regression and a loan pricing model. Report No. 119549 [Internet] Fargo: North Dakota State University, 1988 [cited 2018 Dec 7]. Available from: https://ideas.repec.org/p/ags/nddmrs/119549.html.
- 7. Graham P. Better Bayesian filtering [Internet] PAUL GRAHAM, 2003 [cited 2018 Nov 22]. Available from: http://www.paulgraham.com/better.html.
- 8. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: Pereira F, Burges CJ, Bottou L, Weinberger KQ, eds. Advances in neural information processing systems 25. Red Hook: Curran Associates, Inc, 2012; 1097-105.
- 9. Mnih V, Kavukcuoglu K, Silver D, et al. Human-level control through deep reinforcement learning. Nature 2015; 518: 529-33. ArticlePubMedPDF
- 10. Silver D, Schrittwieser J, Simonyan K, et al. Mastering the game of Go without human knowledge. Nature 2017; 550: 354-9. ArticlePubMedPDF
- 11. Hannun A, Case C, Casper J. Deep speech: scaling up end-to-end speech recognition [Internet] Ithaca: arXiv, Cornell University, 2014 [cited 2018 Nov 22]. Available from: http://arxiv.org/abs/1412.5567.
- 12. Luong MT, Pham H, Manning CD. Effective approaches to attention-based neural machine translation. In: Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing; 2015 Sep 17-21; Lisbon, Portugal. Stroudsburg. Association for Computational Linguistics. 2015; 1412-21. Article
- 13. Wu Y, Schuster M, Chen Z. Google’s neural machine translation system: bridging the gap between human and machine translation [Internet] Ithaca: arXiv, Cornell University, 2016 [cited 2018 Nov 22]. Available from: http://arxiv.org/abs/1609.08144.
- 14. Antol S, Agrawal A, Lu J, et al. VQA: visual question answering. In: Proceedings of the IEEE International Conference on Computer Vision; 2015 Dec 7-13; Santiago, Chile. Washington, DC. IEEE Computer Society. 2015; 2425-33. Article
- 15. Kim JH, Lee SW, Kwak D, et al. Multimodal residual learning for visual QA. In: Lee DD, von Luxburg U, Garnett R, eds. Advances in neural information processing systems 29. Red Hook: NY Curran Associates Inc, 2016; 361-9.
- 16. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE 1998; 86: 2278-324. Article
- 17. Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015 Jun 7-12, Boston, MA, USA. Silver Spring: IEEE Computer Society Press, 2015; 1-9.
- 18. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521: 436-44. ArticlePubMedPDF
- 19. Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997; 9: 1735-80. ArticlePubMed
- 20. Weizenbaum J. ELIZA: a computer program for the study of natural language communication between man and machine. Commun ACM 1966; 9: 36-45. Article
- 21. Shortliffe EH. Mycin: a knowledge-based computer program applied to infectious diseases. In: Proceedings of the Annual Symposium on Computer Application in Medical Care, 1977 Oct 3-5, Washington, DC, USA. New York: Institute of Electrical and Electronics Engineers, 1977; 66-9.
- 22. Heckerman DE, Horvitz EJ, Nathwani BN. Toward normative expert systems: Part I. The Pathfinder project. Methods Inf Med 1992; 31: 90-105. ArticlePubMedPDF
- 23. Heckerman DE, Nathwani BN. Toward normative expert systems: Part II. Probability-based representations for efficient knowledge acquisition and inference. Methods Inf Med 1992; 31: 106-16. ArticlePubMedPDF
- 24. Vyborny CJ, Giger ML. Computer vision and artificial intelligence in mammography. AJR Am J Roentgenol 1994; 162: 699-708. ArticlePubMed
- 25. Kononenko I. Machine learning for medical diagnosis: history, state of the art and perspective. Artif Intell Med 2001; 23: 89-109. ArticlePubMed
- 26. Baker JA, Rosen EL, Lo JY, Gimenez EI, Walsh R, Soo MS. Computeraided detection (CAD) in screening mammography: sensitivity of commercial CAD systems for detecting architectural distortion. AJR Am J Roentgenol 2003; 181: 1083-8. PubMed
- 27. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016; 316: 2402-10. ArticlePubMed
- 28. Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017; 42: 60-88. ArticlePubMed
- 29. Kohli M, Prevedello LM, Filice RW, Geis JR. Implementing machine learning in radiology practice and research. AJR Am J Roentgenol 2017; 208: 754-60. ArticlePubMed
- 30. Shaikhina T, Khovanova NA. Handling limited datasets with neural networks in medical applications: a small-data approach. Artif Intell Med 2017; 75: 51-63. ArticlePubMed
- 31. Angermueller C, Parnamaa T, Parts L, Stegle O. Deep learning for computational biology. Mol Syst Biol 2016; 12: 878.ArticlePubMedPMCPDF
- 32. Torkamani A, Andersen KG, Steinhubl SR, Topol EJ. High-definition medicine. Cell 2017; 170: 828-43. ArticlePubMedPMC
- 33. Wainberg M, Merico D, Delong A, Frey BJ. Deep learning in biomedicine. Nat Biotechnol 2018; 36: 829-38. ArticlePubMedPDF
- 34. Xiong HY, Alipanahi B, Lee LJ, et al. RNA splicing: the human splicing code reveals new insights into the genetic determinants of disease. Science 2015; 347: 1254806.PubMed
- 35. Poplin R, Chang PC, Alexander D, et al. A universal SNP and smallindel variant caller using deep neural networks. Nat Biotechnol 2018; 36: 983-7. ArticlePubMedPDF
- 36. Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med 2018; 1: 18.PubMedPMCPDF
- 37. Fernandes K, Chicco D, Cardoso JS, Fernandes J. Supervised deep learning embeddings for the prediction of cervical cancer diagnosis. PeerJ Comput Sci 2018; 4: e154.ArticlePubMedPMCPDF
- 38. Ye JJ. Artificial intelligence for pathologists is not near: it is here: description of a prototype that can transform how we practice pathology tomorrow. Arch Pathol Lab Med 2015; 139: 929-35. ArticlePubMedPDF
- 39. Beck JR, Salem DN, Estes NA, Pauker SG. A computer-based Markov decision analysis of the management of symptomatic bifascicular block: the threshold probability for pacing. J Am Coll Cardiol 1987; 9: 920-35. ArticlePubMed
- 40. Schaefer AJ, Bailey MD, Shechter SM, Roberts MS. Modeling medical treatment using Markov decision processes. In: Brandeau ML, Sainfort F, Pierskalla WP, eds. Operations research and health care: a handbook of methods and applications. Boston: Kluwer Academic Publisher, 2004; 593-612.
- 41. Alagoz O, Hsu H, Schaefer AJ, Roberts MS. Markov decision processes: a tool for sequential decision making under uncertainty. Med Decis Making 2010; 30: 474-83. ArticlePubMedPDF
- 42. Harbias A, Salmo E, Crump A. Implications of observer variation in Gleason scoring of prostate cancer on clinical management: a collaborative audit. Gulf J Oncolog 2017; 1: 41-5. PubMed
- 43. Ozkan TA, Eruyar AT, Cebeci OO, Memik O, Ozcan L, Kuskonmaz I. Interobserver variability in Gleason histological grading of prostate cancer. Scand J Urol 2016; 50: 420-4. ArticlePubMed
- 44. Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J Pathol Inform 2016; 7: 29.ArticlePubMedPMC
- 45. Komura D, Ishikawa S. Machine learning methods for histopathological image analysis. Comput Struct Biotechnol J 2018; 16: 34-42. ArticlePubMedPMC
- 46. Garud H, Karri SP, Sheet D, et al. High-magnification multi-views based classification of breast fine needle aspiration cytology cell samples using fusion of decisions from deep convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017 Jul 21-26, Honolulu, HI, USA. New York: Institute of Electrical and Electronics Engineers, 2017; 828-33.
- 47. Li Y, Ping W. Cancer metastasis detection with neural conditional random field [Internet] Ithaca: arXiv, Cornell University, 2018 [cited 2018 Nov 22]. Available from: http://arxiv.org/abs/1806.07064.
- 48. Rannen Triki A, Blaschko MB, Jung YM, et al. Intraoperative margin assessment of human breast tissue in optical coherence tomography images using deep neural networks. Comput Med Imaging Graph 2018; 69: 21-32. ArticlePubMed
- 49. Ehteshami Bejnordi B, Mullooly M, Pfeiffer RM, et al. Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod Pathol 2018; 31: 1502-12. ArticlePubMedPMCPDF
- 50. Litjens G, Sánchez CI, Timofeeva N, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep 2016; 6: 26286.ArticlePubMedPMCPDF
- 51. Cires¸an DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Med Image Comput Comput Assist Interv 2013; 16: 411-8. PubMed
- 52. Teramoto A, Tsukamoto T, Kiriyama Y, Fujita H. Automated classification of lung cancer types from cytological images using deep convolutional neural networks. Biomed Res Int 2017; 2017: 4067832.ArticlePubMedPMCPDF
- 53. Yu KH, Zhang C, Berry GJ, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 2016; 7: 12474.ArticlePubMedPMCPDF
- 54. Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018; 24: 1559-67. ArticlePubMedPMCPDF
- 55. Campanella G, Silva VW, Fuchs TJ. Terabyte-scale deep multiple instance learning for classification and localization in pathology [Internet] Ithaca: arXiv, Cornell University, 2018 [cited 2018 Nov 22]. Available from: http://arxiv.org/abs/1805.06983.
- 56. Arvaniti E, Fricker KS, Moret M, et al. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep 2018; 8: 12054.ArticlePubMedPMCPDF
- 57. Zhou N, Fedorov A, Fennessy F, Kikinis R, Gao Y. Large scale digital prostate pathology image analysis combining feature extraction and deep neural network [Internet] Ithaca: arXiv, Cornell University, 2017 [cited 2018 Nov 22]. Available from: http://arxiv.org/abs/1705.02678.
- 58. Nagpal K, Foote D, Liu Y, et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer [Internet] Ithaca: arXiv, Cornell University, 2018 [cited 2018 Nov 22]. Available from: http://arxiv.org/abs/1811.06497.
- 59. Ertosun MG, Rubin DL. Automated grading of gliomas using deep learning in digital pathology images: a modular approach with ensemble of convolutional neural networks. AMIA Annu Symp Proc 2015; 2015: 1899-908. PubMedPMC
- 60. Mobadersany P, Yousefi S, Amgad M, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci U S A 2018; 115: E2970-E9. ArticlePubMedPMC
- 61. Wu M, Yan C, Liu H, Liu Q. Automatic classification of ovarian cancer types from cytological images using deep convolutional neural networks. Biosci Rep 2018; 38: BSR20180289.ArticlePubMedPMCPDF
- 62. Zhang L, Lu L, Nogues I, Summers RM, Liu S, Yao J. DeepPap: deep convolutional networks for cervical cell classification. IEEE J Biomed Health Inform 2017; 21: 1633-43. ArticlePubMed
- 63. Xu M, Papageorgiou DP, Abidi SZ, Dao M, Zhao H, Karniadakis GE. A deep convolutional neural network for classification of red blood cells in sickle cell anemia. PLoS Comput Biol 2017; 13: e1005746. ArticlePubMedPMC
- 64. Meier A, Nekolla K, Earle S, et al. End-to-end learning to predict survival in patients with gastric cancer using convolutional neural networks. Ann Oncol 2018; 29(Suppl 8):mdy269.075. ArticlePDF
- 65. Xie W, Noble JA, Zisserman A. Microscopy cell counting and detection with fully convolutional regression networks. Comput Methods Biomech Biomed Eng Imaging Vis 2016; 6: 283-92. Article
- 66. Tuominen VJ, Ruotoistenmaki S, Viitanen A, Jumppanen M, Isola J. ImmunoRatio: a publicly available web application for quantitative image analysis of estrogen receptor (ER), progesterone receptor (PR), and Ki-67. Breast Cancer Res 2010; 12: R56.ArticlePubMedPMCPDF
- 67. Meijering E. Cell segmentation: 50 years down the road [life sciences]. IEEE Signal Process Mag 2012; 29: 140-5. Article
- 68. Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol 2001; 23: 291-9. PubMed
- 69. Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 1979; 9: 62-6. Article
- 70. Zhang L, Sonka M, Lu L, Summers RM, Yao J. Combining fully convolutional networks and graph-based approach for automated segmentation of cervical cell nuclei. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), 2017 Apr 18-21, Melbourne, VIC, Australia. New York: Institute of Electrical and Electronics Engineers, 2017; 406-9.
- 71. Chen H, Qi X, Yu L, Heng PA. DCAN: deep contour-aware networks for accurate gland segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016 Jun 27-30, Las Vegas, NV, USA. New York: Institute of Electrical and Electronics Engineers, 2016; 2487-96.
- 72. CAMELYON16 Consortium. CAMELYON16. CAMELYON16 ISBI challenge on cancer metastasis detection in lymph node, 2015 [Internet] Grand-Challenges, 2016 [cited 2018 Nov 22]. Available from: https://camelyon16.grand-challenge.org/.
- 73. The Cancer Genome Atlas [Internet] Bethesda: The Cancer Genome Atlas, National Cancer Institute, 2011 [cited 2018 Nov 22]. Available from: https://cancergenome.nih.gov/.
REFERENCES
Figure & Data
References
Citations

- Interpretable Machine Learning Approaches for Identification of Acute Aortic Dissection in Chest Pain Patients
Shuangshuang Li, Kaiwen Zhao, Wen Li, Qingsheng Lu, Jian Zhou, Jia He
Annals of Vascular Surgery.2026; 122: 895. CrossRef - Exploring the status of artificial intelligence for healthcare research in Africa: a bibliometric and thematic analysis
Tabu S. Kondo, Salim A. Diwani, Ally S. Nyamawe, Mohamed M. Mjahidi
AI and Ethics.2025; 5(1): 117. CrossRef - Prioritize Threat Alerts Based on False Positives Qualifiers Provided by Multiple AI Models Using Evolutionary Computation and Reinforcement Learning
Anup Sharma, V. G. Kiran Kumar, Asmita Poojari
Journal of The Institution of Engineers (India): Series B.2025; 106(4): 1305. CrossRef - Artificial intelligence versus human analysis: Interpreting data in elderly fat reduction study
Piotr Sporek, Mariusz Konieczny
Advances in Integrative Medicine.2025; 12(1): 13. CrossRef - Artificial intelligence in healthcare applications targeting cancer diagnosis—part I: data structure, preprocessing and data organization
Anna Luíza Damaceno Araújo, Marcelo Sperandio, Giovanna Calabrese, Sarah S. Faria, Diego Armando Cardona Cardenas, Manoela Domingues Martins, Cristina Saldivia-Siracusa, Daniela Giraldo-Roldán, Caique Mariano Pedroso, Pablo Agustin Vargas, Marcio Ajudarte
Oral Surgery, Oral Medicine, Oral Pathology and Oral Radiology.2025; 140(1): 79. CrossRef - Artificial intelligence–driven digital pathology in urological cancers: current trends and future directions
Inyoung Paik, Geongyu Lee, Joonho Lee, Tae-Yeong Kwak, Hong Koo Ha
Prostate International.2025; 13(4): 181. CrossRef - Optimizing deep learning for accurate blood cell classification: A study on stain normalization and fine-tuning techniques
Mohammed Tareq Mutar, Jaffar Nouri Alalsaidissa, Mustafa Majid Hameed, Ali Almothaffar
Iraqi Journal of Hematology.2025; 14(1): 60. CrossRef - Structural imbalance of medical resources amid population mobility and digital empowerment: a study of national and port-developed provinces in China
Haiwei Fu, Junjie Lu
Frontiers in Public Health.2025;[Epub] CrossRef - Exploring the evolution of artificial intelligence in pathology: a bibliometric and network analysis
Burcu Sanal Yılmaz
Journal of Medicine and Palliative Care.2025; 6(3): 224. CrossRef - ШТУЧНИЙ ІНТЕЛЕКТ У СУЧАСНІЙ СТОМАТОЛОГІЇ
О. І. Бульбук, О. В. Бульбук, О. В. Шутак, Ю. І. Сухоребський
Art of Medicine.2025; : 101. CrossRef - Natural language processing in veterinary pathology: A review
Lev Stimmer, Raoul V. Kuiper, Laura Polledo, Lorenzo Ressel, Josep M. Monné Rodriguez, Inês B. Veiga, Jonathan Williams, Vanessa Herder
Veterinary Pathology.2025; 62(6): 829. CrossRef - Impact of Magnification, Image Type, and Number on Convolutional Neural Network Performance in Differentiating Canine Large Cell Lymphoma From Non‐Lymphoma via Lymph Node Cytology
Christina Pacholec, Hehuang Xie, Julianne Curnin, Amy Lin, Kurt Zimmerman
Veterinary Clinical Pathology.2025;[Epub] CrossRef - Pathology image-based predictive model for individual survival time of early-stage lung adenocarcinoma patients
Vi Thi-Tuong Vo, Hyung-Jeong Yang, Taebum Lee, Soo-Hyung Kim
Scientific Reports.2025;[Epub] CrossRef - Exploring Artificial Intelligence's Potential to Enhance Conventional Anticancer Drug Development
Sorin‐Ștefan Bobolea, Miruna‐Ioana Hinoveanu, Andreea Dimitriu, Miruna‐Andrada Brașoveanu, Cristian‐Nicolae Iliescu, Cristina‐Elena Dinu‐Pîrvu, Mihaela Violeta Ghica, Valentina Anuța, Lăcrămioara Popa, Răzvan Mihai Prisada
Drug Development Research.2025;[Epub] CrossRef - Predicability of PD-L1 expression in cancer cells based solely on H&E-stained sections
Gavino Faa, Matteo Fraschini, Pina Ziranu, Andrea Pretta, Giuseppe Porcu, Luca Saba, Mario Scartozzi, Nazar Shokun, Massimo Rugge
Journal of Pathology Informatics.2025; 19: 100524. CrossRef - Artificial Intelligence in Medicine
Umur Karan, Osman Elbek
Thoracic Research and Practice.2025;[Epub] CrossRef - Whole Slide Imaging Technology and Its Applications: Current and Emerging Perspectives
Ekta Jain, Ankush Patel, Anil V. Parwani, Saba Shafi, Zoya Brar, Shivani Sharma, Sambit K. Mohanty
International Journal of Surgical Pathology.2024; 32(3): 433. CrossRef - ChatGPT as an aid for pathological diagnosis of cancer
Shaivy Malik, Sufian Zaheer
Pathology - Research and Practice.2024; 253: 154989. CrossRef - Computational pathology: A survey review and the way forward
Mahdi S. Hosseini, Babak Ehteshami Bejnordi, Vincent Quoc-Huy Trinh, Lyndon Chan, Danial Hasan, Xingwen Li, Stephen Yang, Taehyo Kim, Haochen Zhang, Theodore Wu, Kajanan Chinniah, Sina Maghsoudlou, Ryan Zhang, Jiadai Zhu, Samir Khaki, Andrei Buin, Fatemeh
Journal of Pathology Informatics.2024; 15: 100357. CrossRef - Applications of artificial intelligence in the field of oral and maxillofacial pathology: a systematic review and meta-analysis
Nishath Sayed Abdul, Ganiga Channaiah Shivakumar, Sunila Bukanakere Sangappa, Marco Di Blasio, Salvatore Crimi, Marco Cicciù, Giuseppe Minervini
BMC Oral Health.2024;[Epub] CrossRef - Machine-learning models are superior to severity scoring systems for the prediction of the mortality of critically ill patients in a tertiary medical center
Ruey-Hsing Chou, Benny Wei-Yun Hsu, Chun-Lin Yu, Tai-Yuan Chen, Shuo-Ming Ou, Kuo-Hua Lee, Vincent S. Tseng, Po-Hsun Huang, Der-Cherng Tarng
Journal of the Chinese Medical Association.2024; 87(4): 369. CrossRef - The Evaluation of Artificial Intelligence Technology for the Differentiation of Fresh Human Blood Cells From Other Species Blood in the Investigation of Crime Scenes
Syed Sajid Hussain Shah, Ekramy Elmorsy, Rashad Qasem Ali Othman, Asmara Syed, Syed Umar Armaghan, Syed Usama Khalid Bokhari, Mahmoud E Elmorsy, Abdulhakim Bawadekji
Cureus.2024;[Epub] CrossRef - A Comparison of Diagnostic and Immunohistochemical Workup and Literature Review Capabilities of Online Artificial Intelligence Assistance Models in Pathology
Johnika Dougan, Netra Patel, Svetoslav Bardarov
Cureus.2024;[Epub] CrossRef - ChatENT: Augmented Large Language Model for Expert Knowledge Retrieval in Otolaryngology–Head and Neck Surgery
Cai Long, Deepak Subburam, Kayle Lowe, André dos Santos, Jessica Zhang, Sang Hwang, Neil Saduka, Yoav Horev, Tao Su, David W.J. Côté, Erin D. Wright
Otolaryngology–Head and Neck Surgery.2024; 171(4): 1042. CrossRef - Artificial intelligence in forensic medicine and related sciences – selected issues = Sztuczna inteligencja w medycynie sądowej i naukach pokrewnych – wybrane zagadnienia
Michał Szeremeta, Julia Janica, Anna Niemcunowicz-Janica
Archives of Forensic Medicine and Criminology.2024; 74(1): 64. CrossRef - Unveiling the landscape of pathomics in personalized immunotherapy for lung cancer: a bibliometric analysis
Lei Yuan, Zhiming Shen, Yibo Shan, Jianwei Zhu, Qi Wang, Yi Lu, Hongcan Shi
Frontiers in Oncology.2024;[Epub] CrossRef - PathEX: Make good choice for whole slide image extraction
Xinda Yang, Ranze Zhang, Yuan Yang, Yu Zhang, Kai Chen, Alberto Marchisio
PLOS ONE.2024; 19(8): e0304702. CrossRef - Automatic point detection on cephalograms using convolutional neural networks: A two-step method
Miki HORI, Makoto JINCHO, Tadasuke HORI, Hironao SEKINE, Akiko KATO, Ken MIYAZAWA, Tatsushi KAWAI
Dental Materials Journal.2024; 43(5): 701. CrossRef - The use of generative artificial intelligence (AI) in teaching and assessment of postgraduate students in pathology and microbiology
Dipmala Das, Asitava Deb Roy, Subhayan Dasgupta, Rohon Das Roy
Indian Journal of Microbiology Research.2024; 11(3): 140. CrossRef - Inteligencia artificial: desafíos éticos y futuros
Jhadson Silva Leonel, Camila Ferreira Silva Leonel, Jonas Byk, Silvania da Conceição Furtado
Revista Bioética.2024;[Epub] CrossRef - Artificial intelligence: ethical and future challenges
Jhadson Silva Leonel, Camila Ferreira Silva Leonel, Jonas Byk, Silvania da Conceição Furtado
Revista Bioética.2024;[Epub] CrossRef - Inteligência artificial: desafios éticos e futuros
Jhadson Silva Leonel, Camila Ferreira Silva Leonel, Jonas Byk, Silvania da Conceição Furtado
Revista Bioética.2024;[Epub] CrossRef - The Constrained-Disorder Principle Assists in Overcoming Significant Challenges in Digital Health: Moving from “Nice to Have” to Mandatory Systems
Noa Hurvitz, Yaron Ilan
Clinics and Practice.2023; 13(4): 994. CrossRef - Building a nonclinical pathology laboratory of the future for pharmaceutical research excellence
D.G. Rudmann, L. Bertrand, A. Zuraw, J. Deiters, M. Staup, Y. Rivenson, J. Kuklyte
Drug Discovery Today.2023; 28(10): 103747. CrossRef - Automated image analysis of keratin 7 staining can predict disease outcome in primary sclerosing cholangitis
Nelli Sjöblom, Sonja Boyd, Anniina Manninen, Sami Blom, Anna Knuuttila, Martti Färkkilä, Johanna Arola
Hepatology Research.2023; 53(4): 322. CrossRef - Application of convolutional neural network for analyzing hepatic fibrosis in mice
Hyun-Ji Kim, Eun Bok Baek, Ji-Hee Hwang, Minyoung Lim, Won Hoon Jung, Myung Ae Bae, Hwa-Young Son, Jae-Woo Cho
Journal of Toxicologic Pathology.2023; 36(1): 21. CrossRef - Machine Learning Techniques for Prognosis Estimation and Knowledge Discovery From Lab Test Results With Application to the COVID-19 Emergency
Alfonso Emilio Gerevini, Roberto Maroldi, Matteo Olivato, Luca Putelli, Ivan Serina
IEEE Access.2023; 11: 83905. CrossRef - Artificial intelligence in dentistry—A review
Hao Ding, Jiamin Wu, Wuyuan Zhao, Jukka P. Matinlinna, Michael F. Burrow, James K. H. Tsoi
Frontiers in Dental Medicine.2023;[Epub] CrossRef - Dental Age Estimation Using the Demirjian Method: Statistical Analysis Using Neural Networks
Byung-Yoon Roh, Jong-Seok Lee, Sang-Beom Lim, Hye-Won Ryu, Su-Jeong Jeon, Ju-Heon Lee, Yo-Seob Seo, Ji-Won Ryu, Jong-Mo Ahn
Korean Journal of Legal Medicine.2023; 47(1): 1. CrossRef - The use of artificial intelligence in health care. Problems of identification of patients' conditions in the processes of detailing the diagnosis
Mintser O
Artificial Intelligence.2023; 28(AI.2023.28): 8. CrossRef - The Effectiveness of Data Augmentation for Mature White Blood Cell Image Classification in Deep Learning — Selection of an Optimal Technique for Hematological Morphology Recognition —
Hiroyuki NOZAKA, Kosuke KAMATA, Kazufumi YAMAGATA
IEICE Transactions on Information and Systems.2023; E106.D(5): 707. CrossRef - Rectal Cancer Stages T2 and T3 Identification Based on Asymptotic Hybrid Feature Maps
Shujing Sun, Jiale Wu, Jian Yao, Yang Cheng, Xin Zhang, Zhihua Lu, Pengjiang Qian
Computer Modeling in Engineering & Sciences.2023; 137(1): 923. CrossRef - How to use AI in pathology
Peter Schüffler, Katja Steiger, Wilko Weichert
Genes, Chromosomes and Cancer.2023; 62(9): 564. CrossRef - Cutting-Edge Technologies for Digital Therapeutics: A Review and Architecture Proposals for Future Directions
Joo Hun Yoo, Harim Jeong, Tai-Myoung Chung
Applied Sciences.2023; 13(12): 6929. CrossRef - A convolutional neural network STIFMap reveals associations between stromal stiffness and EMT in breast cancer
Connor Stashko, Mary-Kate Hayward, Jason J. Northey, Neil Pearson, Alastair J. Ironside, Johnathon N. Lakins, Roger Oria, Marie-Anne Goyette, Lakyn Mayo, Hege G. Russnes, E. Shelley Hwang, Matthew L. Kutys, Kornelia Polyak, Valerie M. Weaver
Nature Communications.2023;[Epub] CrossRef - Artificial Intelligence-Based PTEN Loss Assessment as an Early Predictor of Prostate Cancer Metastasis After Surgery: A Multicenter Retrospective Study
Palak Patel, Stephanie Harmon, Rachael Iseman, Olga Ludkowski, Heidi Auman, Sarah Hawley, Lisa F. Newcomb, Daniel W. Lin, Peter S. Nelson, Ziding Feng, Hilary D. Boyer, Maria S. Tretiakova, Larry D. True, Funda Vakar-Lopez, Peter R. Carroll, Matthew R. Co
Modern Pathology.2023; 36(10): 100241. CrossRef - Minimum resolution requirements of digital pathology images for accurate classification
Lydia Neary-Zajiczek, Linas Beresna, Benjamin Razavi, Vijay Pawar, Michael Shaw, Danail Stoyanov
Medical Image Analysis.2023; 89: 102891. CrossRef - Artificial Intelligence in the Pathology of Gastric Cancer
Sangjoon Choi, Seokhwi Kim
Journal of Gastric Cancer.2023; 23(3): 410. CrossRef - Endoscopic Ultrasound-Based Artificial Intelligence Diagnosis of Pancreatic Cystic Neoplasms
Jin-Seok Park, Seok Jeong
The Korean Journal of Pancreas and Biliary Tract.2023; 28(3): 53. CrossRef - Framework for Classifying Explainable Artificial Intelligence (XAI) Algorithms in Clinical Medicine
Thomas Gniadek, Jason Kang, Talent Theparee, Jacob Krive
Online Journal of Public Health Informatics.2023; 15: e50934. CrossRef - A Literature Review of the Future of Oral Medicine and Radiology, Oral Pathology, and Oral Surgery in the Hands of Technology
Ishita Singhal, Geetpriya Kaur, Dirk Neefs, Aparna Pathak
Cureus.2023;[Epub] CrossRef - AI-Powered Biomolecular-Specific and Label-Free Multispectral Imaging Rapidly Detects Malignant Neoplasm in Surgically Excised Breast Tissue Specimens
Rishikesh Pandey, David Fournier, Gary Root, Machele Riccio, Aditya Shirvalkar, Gianfranco Zamora, Noel Daigneault, Michael Sapack, Minghao Zhong, Malini Harigopal
Archives of Pathology & Laboratory Medicine.2023; 147(11): 1298. CrossRef - Artificial intelligence for patient scheduling in the real-world health care setting: A metanarrative review
Dacre R.T. Knight, Christopher A. Aakre, Christopher V. Anstine, Bala Munipalli, Parisa Biazar, Ghada Mitri, Jose Raul Valery, Tara Brigham, Shehzad K. Niazi, Adam I. Perlman, John D. Halamka, Abd Moain Abu Dabrh
Health Policy and Technology.2023; 12(4): 100824. CrossRef - Towards Autonomous Healthcare: Integrating Artificial Intelligence (AI) for Personalized Medicine and Disease Prediction
Nitin Rane, Saurabh Choudhary, Jayesh Rane
SSRN Electronic Journal.2023;[Epub] CrossRef - Medical imaging and multimodal artificial intelligence models for streamlining and enhancing cancer care: opportunities and challenges
Kevin Pierre, Manas Gupta, Abheek Raviprasad, Seyedeh Mehrsa Sadat Razavi, Anjali Patel, Keith Peters, Bruno Hochhegger, Anthony Mancuso, Reza Forghani
Expert Review of Anticancer Therapy.2023; 23(12): 1265. CrossRef - Automated differential diagnostics of respiratory diseases using an electronic stethoscope
Diana Arhypenko, Denis Panaskin, Dmytro Babko
Polish Journal of Medical Physics and Engineering.2023; 29(4): 208. CrossRef - Application of machine learning in identification of pathogenic microbes
Lakshmi Venkata S Kutikuppala, Kanishk K Adhit, Reewen George D Silva
Digital Medicine.2023;[Epub] CrossRef - The Beginning of a New Era
C Nandini, Shaik Basha, Aarchi Agarawal, R Parikh Neelampari, Krishna P Miyapuram, R Jadeja Nileshwariba
Advances in Human Biology.2023; 13(1): 4. CrossRef - Artificial Intelligence in Respiratory Medicine
K Kalaiyarasan, R Sridhar
Journal of Association of Pulmonologist of Tamil Nadu.2023; 6(2): 53. CrossRef - Automated abstraction of myocardial perfusion imaging reports using natural language processing
Parija Sharedalal, Ajay Singh, Neal Shah, Diwakar Jain
Journal of Nuclear Cardiology.2022; 29(3): 1188. CrossRef - Polyploid giant cancer cell characterization: New frontiers in predicting response to chemotherapy in breast cancer
Geetanjali Saini, Shriya Joshi, Chakravarthy Garlapati, Hongxiao Li, Jun Kong, Jayashree Krishnamurthy, Michelle D. Reid, Ritu Aneja
Seminars in Cancer Biology.2022; 81: 220. CrossRef - A Comprehensive Review of Markov Random Field and Conditional Random Field Approaches in Pathology Image Analysis
Yixin Li, Chen Li, Xiaoyan Li, Kai Wang, Md Mamunur Rahaman, Changhao Sun, Hao Chen, Xinran Wu, Hong Zhang, Qian Wang
Archives of Computational Methods in Engineering.2022; 29(1): 609. CrossRef - Artificial intelligence in oncology: From bench to clinic
Jamal Elkhader, Olivier Elemento
Seminars in Cancer Biology.2022; 84: 113. CrossRef - Yeast‐like organisms phagocytosed by circulating neutrophils: Evidence of disseminated histoplasmosis
Yue Zhao, Jenna McCracken, Endi Wang
International Journal of Laboratory Hematology.2022; 44(1): 51. CrossRef - Whole-slide imaging, tissue image analysis, and artificial intelligence in veterinary pathology: An updated introduction and review
Aleksandra Zuraw, Famke Aeffner
Veterinary Pathology.2022; 59(1): 6. CrossRef - A comprehensive review of computer-aided whole-slide image analysis: from datasets to feature extraction, segmentation, classification and detection approaches
Xintong Li, Chen Li, Md Mamunur Rahaman, Hongzan Sun, Xiaoqi Li, Jian Wu, Yudong Yao, Marcin Grzegorzek
Artificial Intelligence Review.2022; 55(6): 4809. CrossRef - Liquid Biopsy and Artificial Intelligence as Tools to Detect Signatures of Colorectal Malignancies: A Modern Approach in Patient’s Stratification
Octav Ginghina, Ariana Hudita, Marius Zamfir, Andrada Spanu, Mara Mardare, Irina Bondoc, Laura Buburuzan, Sergiu Emil Georgescu, Marieta Costache, Carolina Negrei, Cornelia Nitipir, Bianca Galateanu
Frontiers in Oncology.2022;[Epub] CrossRef - Automated bone marrow cytology using deep learning to generate a histogram of cell types
Rohollah Moosavi Tayebi, Youqing Mu, Taher Dehkharghanian, Catherine Ross, Monalisa Sur, Ronan Foley, Hamid R. Tizhoosh, Clinton J. V. Campbell
Communications Medicine.2022;[Epub] CrossRef - Risultati di esami di laboratorio per intelligenza artificiale e "machine learning"
Marco PRADELLA
La Rivista Italiana della Medicina di Laboratorio.2022;[Epub] CrossRef - The Deception of Certainty: how Non-Interpretable Machine Learning Outcomes Challenge the Epistemic Authority of Physicians. A deliberative-relational Approach
Florian Funer
Medicine, Health Care and Philosophy.2022; 25(2): 167. CrossRef - Deep discriminative learning model with calibrated attention map for the automated diagnosis of diffuse large B-cell lymphoma
Sautami Basu, Ravinder Agarwal, Vishal Srivastava
Biomedical Signal Processing and Control.2022; 76: 103728. CrossRef - Question and Answer Techniques for Financial Audits in Universities Based on Deep Learning
Qiang Li, Hangjun Che
Mathematical Problems in Engineering.2022; 2022: 1. CrossRef - Noninvasive Screening Tool for Hyperkalemia Using a Single-Lead Electrocardiogram and Deep Learning: Development and Usability Study
Erdenebayar Urtnasan, Jung Hun Lee, Byungjin Moon, Hee Young Lee, Kyuhee Lee, Hyun Youk
JMIR Medical Informatics.2022; 10(6): e34724. CrossRef - Impact of artificial intelligence on pathologists’ decisions: an experiment
Julien Meyer, April Khademi, Bernard Têtu, Wencui Han, Pria Nippak, David Remisch
Journal of the American Medical Informatics Association.2022; 29(10): 1688. CrossRef - Rapid Screening Using Pathomorphologic Interpretation to Detect BRAFV600E Mutation and Microsatellite Instability in Colorectal Cancer
Satoshi Fujii, Daisuke Kotani, Masahiro Hattori, Masato Nishihara, Toshihide Shikanai, Junji Hashimoto, Yuki Hama, Takuya Nishino, Mizuto Suzuki, Ayatoshi Yoshidumi, Makoto Ueno, Yoshito Komatsu, Toshiki Masuishi, Hiroki Hara, Taito Esaki, Yoshiaki Nakamu
Clinical Cancer Research.2022; 28(12): 2623. CrossRef - Using Deep Learning to Predict Final HER2 Status in Invasive Breast Cancers That are Equivocal (2+) by Immunohistochemistry
Sean A. Rasmussen, Valerie J. Taylor, Alexi P. Surette, Penny J. Barnes, Gillian C. Bethune
Applied Immunohistochemistry & Molecular Morphology.2022; 30(10): 668. CrossRef - Deep Neural Network for the Prediction of KRAS Genotype in Rectal Cancer
Waleed M Ghareeb, Eman Draz, Khaled Madbouly, Ahmed H Hussein, Mohammed Faisal, Wagdi Elkashef, Mona Hany Emile, Marcus Edelhamre, Seon Hahn Kim, Sameh Hany Emile
Journal of the American College of Surgeons.2022; 235(3): 482. CrossRef - Next Generation Digital Pathology: Emerging Trends and Measurement Challenges for Molecular Pathology
Alex Dexter, Dimitrios Tsikritsis, Natalie A. Belsey, Spencer A. Thomas, Jenny Venton, Josephine Bunch, Marina Romanchikova
Journal of Molecular Pathology.2022; 3(3): 168. CrossRef - Animation Design of Multisensor Data Fusion Based on Optimized AVOD Algorithm
Li Ding, Guobing Wei, Kai Zhang, Gengxin Sun
Journal of Sensors.2022; 2022: 1. CrossRef - Study on Machine Translation Teaching Model Based on Translation Parallel Corpus and Exploitation for Multimedia Asian Information Processing
Yan Gong
ACM Transactions on Asian and Low-Resource Language Information Processing.2022;[Epub] CrossRef - Analysis and Estimation of Pathological Data and Findings with Deep Learning Methods
Ahmet Anıl ŞAKIR, Ali Hakan IŞIK, Özlem ÖZMEN, Volkan İPEK
Veterinary Journal of Mehmet Akif Ersoy University.2022; 7(3): 175. CrossRef - Artificial Intelligence in Pathology: Friend or Enemy?
Selim Sevim, Ezgi Dicle Serbes, Murat Bahadır, Mustafa Said Kartal, Serpil Dizbay Sak
Journal of Ankara University Faculty of Medicine.2022; 75(1): 13. CrossRef - Assessment of knowledge, attitude, and practice regarding artificial intelligence in histopathology
M. Indu, Vidya Gurram Shankar, Latha Mary Cherian, Revathi Krishna, Sabu Paul, Pradeesh Sathyan
Saudi Journal of Oral Sciences.2022; 9(3): 157. CrossRef - Evaluation Challenges in the Validation of B7-H3 as Oral Tongue Cancer Prognosticator
Meri Sieviläinen, Anna Maria Wirsing, Aini Hyytiäinen, Rabeia Almahmoudi, Priscila Rodrigues, Inger-Heidi Bjerkli, Pirjo Åström, Sanna Toppila-Salmi, Timo Paavonen, Ricardo D. Coletta, Elin Hadler-Olsen, Tuula Salo, Ahmed Al-Samadi
Head and Neck Pathology.2021; 15(2): 469. CrossRef - Amsterdam International Consensus Meeting: tumor response scoring in the pathology assessment of resected pancreatic cancer after neoadjuvant therapy
Boris V. Janssen, Faik Tutucu, Stijn van Roessel, Volkan Adsay, Olca Basturk, Fiona Campbell, Claudio Doglioni, Irene Esposito, Roger Feakins, Noriyoshi Fukushima, Anthony J. Gill, Ralph H. Hruban, Jeffrey Kaplan, Bas Groot Koerkamp, Seung-Mo Hong, Alyssa
Modern Pathology.2021; 34(1): 4. CrossRef - Fabrication of ultra-thin 2D covalent organic framework nanosheets and their application in functional electronic devices
Weikang Wang, Weiwei Zhao, Haotian Xu, Shujuan Liu, Wei Huang, Qiang Zhao
Coordination Chemistry Reviews.2021; 429: 213616. CrossRef - Generalizability of Deep Learning System for the Pathologic Diagnosis of Various Cancers
Hyun-Jong Jang, In Hye Song, Sung Hak Lee
Applied Sciences.2021; 11(2): 808. CrossRef - Integrated digital pathology at scale: A solution for clinical diagnostics and cancer research at a large academic medical center
Peter J Schüffler, Luke Geneslaw, D Vijay K Yarlagadda, Matthew G Hanna, Jennifer Samboy, Evangelos Stamelos, Chad Vanderbilt, John Philip, Marc-Henri Jean, Lorraine Corsale, Allyne Manzo, Neeraj H G Paramasivam, John S Ziegler, Jianjiong Gao, Juan C Peri
Journal of the American Medical Informatics Association.2021; 28(9): 1874. CrossRef - Translational Applications of Artificial Intelligence and Machine Learning for Diagnostic Pathology in Lymphoid Neoplasms: A Comprehensive and Evolutive Analysis
Julia Moran-Sanchez, Antonio Santisteban-Espejo, Miguel Angel Martin-Piedra, Jose Perez-Requena, Marcial Garcia-Rojo
Biomolecules.2021; 11(6): 793. CrossRef - Development and operation of a digital platform for sharing pathology image data
Yunsook Kang, Yoo Jung Kim, Seongkeun Park, Gun Ro, Choyeon Hong, Hyungjoon Jang, Sungduk Cho, Won Jae Hong, Dong Un Kang, Jonghoon Chun, Kyoungbun Lee, Gyeong Hoon Kang, Kyoung Chul Moon, Gheeyoung Choe, Kyu Sang Lee, Jeong Hwan Park, Won-Ki Jeong, Se Yo
BMC Medical Informatics and Decision Making.2021;[Epub] CrossRef - Sliding window based deep ensemble system for breast cancer classification
Amin Alqudah, Ali Mohammad Alqudah
Journal of Medical Engineering & Technology.2021; 45(4): 313. CrossRef - Artificial intelligence and computational pathology
Miao Cui, David Y. Zhang
Laboratory Investigation.2021; 101(4): 412. CrossRef - Effects of Image Quantity and Image Source Variation on Machine Learning Histology Differential Diagnosis Models
Elham Vali-Betts, Kevin J. Krause, Alanna Dubrovsky, Kristin Olson, John Paul Graff, Anupam Mitra, Ananya Datta-Mitra, Kenneth Beck, Aristotelis Tsirigos, Cynthia Loomis, Antonio Galvao Neto, Esther Adler, Hooman H. Rashidi
Journal of Pathology Informatics.2021; 12(1): 5. CrossRef - Feasibility of deep learning‐based fully automated classification of microsatellite instability in tissue slides of colorectal cancer
Sung Hak Lee, In Hye Song, Hyun‐Jong Jang
International Journal of Cancer.2021; 149(3): 728. CrossRef - Artificial intelligence in healthcare
Yamini D Shah, Shailvi M Soni, Manish P Patel
Indian Journal of Pharmacy and Pharmacology.2021; 8(2): 102. CrossRef - Proof of Concept for a Deep Learning Algorithm for Identification and Quantification of Key Microscopic Features in the Murine Model of DSS-Induced Colitis
Agathe Bédard, Thomas Westerling-Bui, Aleksandra Zuraw
Toxicologic Pathology.2021; 49(4): 897. CrossRef - An empirical analysis of machine learning frameworks for digital pathology in medical science
S.K.B. Sangeetha, R Dhaya, Dhruv T Shah, R Dharanidharan, K. Praneeth Sai Reddy
Journal of Physics: Conference Series.2021; 1767(1): 012031. CrossRef - Application of Single-Cell Approaches to Study Myeloproliferative Neoplasm Biology
Daniel Royston, Adam J. Mead, Bethan Psaila
Hematology/Oncology Clinics of North America.2021; 35(2): 279. CrossRef - Idiosyncratic Drug-Induced Liver Injury (DILI) and Herb-Induced Liver Injury (HILI): Diagnostic Algorithm Based on the Quantitative Roussel Uclaf Causality Assessment Method (RUCAM)
Rolf Teschke, Gaby Danan
Diagnostics.2021; 11(3): 458. CrossRef - Searching Images for Consensus
Hamid R. Tizhoosh, Phedias Diamandis, Clinton J.V. Campbell, Amir Safarpoor, Shivam Kalra, Danial Maleki, Abtin Riasatian, Morteza Babaie
The American Journal of Pathology.2021; 191(10): 1702. CrossRef - Automated Classification and Segmentation in Colorectal Images Based on Self‐Paced Transfer Network
Yao Yao, Shuiping Gou, Ru Tian, Xiangrong Zhang, Shuixiang He, Zhiguo Zhou
BioMed Research International.2021;[Epub] CrossRef - Artificial intelligence and sleep: Advancing sleep medicine
Nathaniel F. Watson, Christopher R. Fernandez
Sleep Medicine Reviews.2021; 59: 101512. CrossRef - Prospective Of Artificial Intelligence: Emerging Trends In Modern Biosciences Research
Pradeep Kumar, Ajit Kumar Singh Yadav, Abhishek Singh
IOP Conference Series: Materials Science and Engineering.2021; 1020(1): 012008. CrossRef - Use and Control of Artificial Intelligence in Patients Across the Medical Workflow: Single-Center Questionnaire Study of Patient Perspectives
Simon Lennartz, Thomas Dratsch, David Zopfs, Thorsten Persigehl, David Maintz, Nils Große Hokamp, Daniel Pinto dos Santos
Journal of Medical Internet Research.2021; 23(2): e24221. CrossRef - HEAL: an automated deep learning framework for cancer histopathology image analysis
Yanan Wang, Nicolas Coudray, Yun Zhao, Fuyi Li, Changyuan Hu, Yao-Zhong Zhang, Seiya Imoto, Aristotelis Tsirigos, Geoffrey I Webb, Roger J Daly, Jiangning Song, Zhiyong Lu
Bioinformatics.2021; 37(22): 4291. CrossRef - A Review of Applications of Artificial Intelligence in Gastroenterology
Khalid Nawab, Ravi Athwani, Awais Naeem, Muhammad Hamayun, Momna Wazir
Cureus.2021;[Epub] CrossRef - Evaluating Cancer-Related Biomarkers Based on Pathological Images: A Systematic Review
Xiaoliang Xie, Xulin Wang, Yuebin Liang, Jingya Yang, Yan Wu, Li Li, Xin Sun, Pingping Bing, Binsheng He, Geng Tian, Xiaoli Shi
Frontiers in Oncology.2021;[Epub] CrossRef - Deep learning-based histopathological segmentation for whole slide images of colorectal cancer in a compressed domain
Hyeongsub Kim, Hongjoon Yoon, Nishant Thakur, Gyoyeon Hwang, Eun Jung Lee, Chulhong Kim, Yosep Chong
Scientific Reports.2021;[Epub] CrossRef - Deep Learning on Oral Squamous Cell Carcinoma Ex Vivo Fluorescent Confocal Microscopy Data: A Feasibility Study
Veronika Shavlokhova, Sameena Sandhu, Christa Flechtenmacher, Istvan Koveshazi, Florian Neumeier, Víctor Padrón-Laso, Žan Jonke, Babak Saravi, Michael Vollmer, Andreas Vollmer, Jürgen Hoffmann, Michael Engel, Oliver Ristow, Christian Freudlsperger
Journal of Clinical Medicine.2021; 10(22): 5326. CrossRef - A Pathologist-Annotated Dataset for Validating Artificial Intelligence: A Project Description and Pilot Study
Sarah N. Dudgeon, Si Wen, Matthew G. Hanna, Rajarsi Gupta, Mohamed Amgad, Manasi Sheth, Hetal Marble, Richard Huang, Markus D. Herrmann, Clifford H. Szu, Darick Tong, Bruce Werness, Evan Szu, Denis Larsimont, Anant Madabhushi, Evangelos Hytopoulos, Weijie
Journal of Pathology Informatics.2021; 12(1): 45. CrossRef - Artificial Intelligence in Medicine: A Multinational Multi-Center Survey on the Medical and Dental Students' Perception
Sotirios Bisdas, Constantin-Cristian Topriceanu, Zosia Zakrzewska, Alexandra-Valentina Irimia, Loizos Shakallis, Jithu Subhash, Maria-Madalina Casapu, Jose Leon-Rojas, Daniel Pinto dos Santos, Dilys Miriam Andrews, Claudia Zeicu, Ahmad Mohammad Bouhuwaish
Frontiers in Public Health.2021;[Epub] CrossRef - Digital/Computational Technology for Molecular Cytology Testing: A Short Technical Note with Literature Review
Robert Y. Osamura, Naruaki Matsui, Masato Kawashima, Hiroyasu Saiga, Maki Ogura, Tomoharu Kiyuna
Acta Cytologica.2021; 65(4): 342. CrossRef - Advances in Digital Pathology: From Artificial Intelligence to Label-Free Imaging
Frederik Großerueschkamp, Hendrik Jütte, Klaus Gerwert, Andrea Tannapfel
Visceral Medicine.2021; 37(6): 482. CrossRef - Feasibility of fully automated classification of whole slide images based on deep learning
Kyung-Ok Cho, Sung Hak Lee, Hyun-Jong Jang
The Korean Journal of Physiology & Pharmacology.2020; 24(1): 89. CrossRef - Same same but different: A Web‐based deep learning application revealed classifying features for the histopathologic distinction of cortical malformations
Joshua Kubach, Angelika Muhlebner‐Fahrngruber, Figen Soylemezoglu, Hajime Miyata, Pitt Niehusmann, Mrinalini Honavar, Fabio Rogerio, Se‐Hoon Kim, Eleonora Aronica, Rita Garbelli, Samuel Vilz, Alexander Popp, Stefan Walcher, Christoph Neuner, Michael Schol
Epilepsia.2020; 61(3): 421. CrossRef - Segmentation and Classification in Digital Pathology for Glioma Research: Challenges and Deep Learning Approaches
Tahsin Kurc, Spyridon Bakas, Xuhua Ren, Aditya Bagari, Alexandre Momeni, Yue Huang, Lichi Zhang, Ashish Kumar, Marc Thibault, Qi Qi, Qian Wang, Avinash Kori, Olivier Gevaert, Yunlong Zhang, Dinggang Shen, Mahendra Khened, Xinghao Ding, Ganapathy Krishnamu
Frontiers in Neuroscience.2020;[Epub] CrossRef - Artificial intelligence as the next step towards precision pathology
B. Acs, M. Rantalainen, J. Hartman
Journal of Internal Medicine.2020; 288(1): 62. CrossRef - Introduction to digital pathology and computer-aided pathology
Soojeong Nam, Yosep Chong, Chan Kwon Jung, Tae-Yeong Kwak, Ji Youl Lee, Jihwan Park, Mi Jung Rho, Heounjeong Go
Journal of Pathology and Translational Medicine.2020; 54(2): 125. CrossRef - Artificial intelligence with multi-functional machine learning platform development for better healthcare and precision medicine
Zeeshan Ahmed, Khalid Mohamed, Saman Zeeshan, XinQi Dong
Database.2020;[Epub] CrossRef - Scoring pleurisy in slaughtered pigs using convolutional neural networks
Abigail R. Trachtman, Luca Bergamini, Andrea Palazzi, Angelo Porrello, Andrea Capobianco Dondona, Ercole Del Negro, Andrea Paolini, Giorgio Vignola, Simone Calderara, Giuseppe Marruchella
Veterinary Research.2020;[Epub] CrossRef - Current Status of Computational Intelligence Applications in Dermatological Clinical Practice
Carmen Rodríguez-Cerdeira, José Luís González-Cespón, Roberto Arenas
The Open Dermatology Journal.2020; 14(1): 6. CrossRef - New unified insights on deep learning in radiological and pathological images: Beyond quantitative performances to qualitative interpretation
Yoichi Hayashi
Informatics in Medicine Unlocked.2020; 19: 100329. CrossRef - Artificial Intelligence in Cardiology: Present and Future
Francisco Lopez-Jimenez, Zachi Attia, Adelaide M. Arruda-Olson, Rickey Carter, Panithaya Chareonthaitawee, Hayan Jouni, Suraj Kapa, Amir Lerman, Christina Luong, Jose R. Medina-Inojosa, Peter A. Noseworthy, Patricia A. Pellikka, Margaret M. Redfield, Vero
Mayo Clinic Proceedings.2020; 95(5): 1015. CrossRef - Artificial intelligence in oncology
Hideyuki Shimizu, Keiichi I. Nakayama
Cancer Science.2020; 111(5): 1452. CrossRef - Artificial intelligence and the future of global health
Nina Schwalbe, Brian Wahl
The Lancet.2020; 395(10236): 1579. CrossRef - The future of pathology is digital
J.D. Pallua, A. Brunner, B. Zelger, M. Schirmer, J. Haybaeck
Pathology - Research and Practice.2020; 216(9): 153040. CrossRef - Weakly-supervised learning for lung carcinoma classification using deep learning
Fahdi Kanavati, Gouji Toyokawa, Seiya Momosaki, Michael Rambeau, Yuka Kozuma, Fumihiro Shoji, Koji Yamazaki, Sadanori Takeo, Osamu Iizuka, Masayuki Tsuneki
Scientific Reports.2020;[Epub] CrossRef - The use of artificial intelligence, machine learning and deep learning in oncologic histopathology
Ahmed S. Sultan, Mohamed A. Elgharib, Tiffany Tavares, Maryam Jessri, John R. Basile
Journal of Oral Pathology & Medicine.2020; 49(9): 849. CrossRef - Convergence of Digital Pathology and Artificial Intelligence Tools in Anatomic Pathology Practice: Current Landscape and Future Directions
Anil V. Parwani, Mahul B. Amin
Advances in Anatomic Pathology.2020; 27(4): 221. CrossRef - Advances in tissue-based imaging: impact on oncology research and clinical practice
Arman Rahman, Chowdhury Jahangir, Seodhna M. Lynch, Nebras Alattar, Claudia Aura, Niamh Russell, Fiona Lanigan, William M. Gallagher
Expert Review of Molecular Diagnostics.2020; 20(10): 1027. CrossRef - Current Trends of Artificial Intelligence for Colorectal Cancer Pathology Image Analysis: A Systematic Review
Nishant Thakur, Hongjun Yoon, Yosep Chong
Cancers.2020; 12(7): 1884. CrossRef - Explainable Machine Learning Model for Predicting GI Bleed Mortality in the Intensive Care Unit
Farah Deshmukh, Shamel S. Merchant
American Journal of Gastroenterology.2020; 115(10): 1657. CrossRef - Prediction of clinically actionable genetic alterations from colorectal cancer histopathology images using deep learning
Hyun-Jong Jang, Ahwon Lee, J Kang, In Hye Song, Sung Hak Lee
World Journal of Gastroenterology.2020; 26(40): 6207. CrossRef - Application of system analysis methods for modeling the development of hand-arm vibration syndrome: problems and approaches to solution
M P Diakovich, M V Krivov
Journal of Physics: Conference Series.2020; 1661(1): 012029. CrossRef - Histo-ELISA technique for quantification and localization of tissue components
Zhongmin Li, Silvia Goebel, Andreas Reimann, Martin Ungerer
Scientific Reports.2020;[Epub] CrossRef - Role of artificial intelligence in diagnostic oral pathology-A modern approach
Ayinampudi Bhargavi Krishna, Azra Tanveer, Pancha Venkat Bhagirath, Ashalata Gannepalli
Journal of Oral and Maxillofacial Pathology.2020; 24(1): 152. CrossRef - Applications of deep learning for the analysis of medical data
Hyun-Jong Jang, Kyung-Ok Cho
Archives of Pharmacal Research.2019; 42(6): 492. CrossRef - PROMISE CLIP Project: A Retrospective, Multicenter Study for Prostate Cancer that Integrates Clinical, Imaging and Pathology Data
Jihwan Park, Mi Jung Rho, Yong Hyun Park, Chan Kwon Jung, Yosep Chong, Choung-Soo Kim, Heounjeong Go, Seong Soo Jeon, Minyong Kang, Hak Jong Lee, Sung Il Hwang, Ji Youl Lee
Applied Sciences.2019; 9(15): 2982. CrossRef - Key challenges for delivering clinical impact with artificial intelligence
Christopher J. Kelly, Alan Karthikesalingam, Mustafa Suleyman, Greg Corrado, Dominic King
BMC Medicine.2019;[Epub] CrossRef - Deep Learning for Whole Slide Image Analysis: An Overview
Neofytos Dimitriou, Ognjen Arandjelović, Peter D. Caie
Frontiers in Medicine.2019;[Epub] CrossRef - Barriers to Artificial Intelligence Adoption in Healthcare Management: A Systematic Review
Mir Mohammed Assadullah
SSRN Electronic Journal .2019;[Epub] CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure



Fig. 1.
Fig. 2.
Fig. 3.
| Term | Abbreviation | Explanation |
|---|---|---|
| Artificial intelligence | AI | Intelligence represented by artificial things |
| Machine learning | ML | Data-driven statistical learning approach to AI |
| Deep learning | DL | Deep neural network based ML |
| Convolutional neural network | CNN | Neural network suitable for data with locality, e.g. image |
| Recurrent neural network | RNN | Neural network suitable for data with order dependency, e.g. sentence |
| Long short-term memory | LSTM | Recurrent neuron suitable for learning long-term dependency |
| Support vector machine | SVM | ML method that separates with regard to the trained hyperplane |
| k-nearest neighbor (search) | k-NN | ML method that classifies based on the classes of k similar training data |
| Conditional random field | CRF | ML method suitable for data with spatial/temporal dependency |
| Markov decision process | MDP | Modeling framework for a series of decisions and resulting outcomes |
| Multiple instance learning | MIL | ML approach suitable for labeled sets (whole slides) of unlabeled instances (lesions) |
| Region-of-interest | ROI | Image region containing things of predefined interest, e.g. nuclei, stroma, etc. |
| Area under receiver operating characteristic curve | AUC | Performance measure based on the area under the receiver operating characteristic curve, varying from 0.5 (lowest) to 1.0 (highest) |
| Author (year) | Disease | Data | Task | Model | Augmentation | Performance |
|---|---|---|---|---|---|---|
| Garud et al. (2017) [46] | Breast cancer | FNA cytology/175 (images) | Decision Benign/cancer | CNN | None | Image level decision acc. 89.7% |
| Li and Ping (2018) [47] | Lymph node metastasis | CAMELYON16/400 (WSIs) | Decision Yes/no | CNN + CRF | Color jitter, rotation, etc. | Patch level decision acc. 93.8% |
| Rannen Triki et al. (2018) [48] | Breast cancer | Frozen section OCT/4,921 (frames) | Decision Benign/cancer | CNN | None | Patch level decision acc. 94.96% |
| Ehteshami Bejnordi et al. (2018) [49] | Breast cancer | BREAST Stamp/2,387 (WSIs) | Decision Benign/cancer | CNN + CNN | None | WSI level decision AUC 0.962 |
| Litjens et al. (2016) [50] | Lymph node metastasis | Lymph node specimen/271 (samples) | Decision Yes/no | CNN | None | Sample level decision AUC 0.90 |
| Cires¸ an et al. (2013) [51] | Breast cancer | MITOS/300 mitosis in 50 images | Mitosis detection | CNN | Rotation, flip, etc. | Detection F1-score 0.782 |
| Teramoto et al. (2017) [52] | Lung cancer | FNA cytology/298 (images) | Classification | CNN | Rotation, flip, etc. | Overall classification acc. 71.1% |
| Adeno-Squamous cell | ||||||
| Small cell | ||||||
| Yu et al. (2016) [53] | Lung cancer | TCGA-LUAD/1,074 | Decision Benign/cancer | SVM | None | Patch level decision AUC 0.85 |
| TCGA-LUSC/1,111 | Survival analysis | |||||
| Stanford TMA/294 (samples) | ||||||
| Coudray et al. (2018) [54] | Lung cancer | TCGA lung cancer/1,635 (samples) | Classification | CNN | None | Overall classification AUC 0.97 |
| Adeno-Squamous cell | STK11 mutation decision AUC 0.85 | |||||
| Benign | ||||||
| Multi-task decision | ||||||
| Gene mutation | ||||||
| Campanella et al. (2018) [55] | Prostate cancer | Needle biopsy/12,160 (samples) | Decision Benign/cancer | CNN (MIL) | None | Sample level decision AUC 0.979 |
| Arvaniti et al. (2018) [56] | Prostate cancer | TMA/886 (samples) | Classification Gleason score | CNN +scoring rule | Rotation, flip, color jitter | Model-pathologist Cohen’s kappa 0.71 |
| Zhou et al. (2017) [57] | Prostate cancer | TCGA-PRAD/368 (cases) | Decision 3 + 4/4 + 3 | CNN | None | Sample level decision acc. 75% |
| Nagpal et al. (2018) [58] | Prostate cancer | TCGA-PRAD + others/train 1,226, test 331 (slides) | Classification Gleason group | CNN + k-NN | None | Overall classification acc. 70% |
| Survival analysis | C-index 0.697 | |||||
| Litjens et al. (2016) [50] | Prostate cancer | Needle biopsy / 225 (WSIs) | Decision Benign/cancer | CNN | None | Slide level decision AUC 0.99 |
| Ertosun and Rubin (2015) [59] | Brain cancer | TCGA-GBM & LGG (unknown size) | Classification | CNN + CNN | Color transform to H&E | GBM/LGG decision acc. 96% |
| GBM | LGG grade decision acc. 71% | |||||
| LGG grade 2 | ||||||
| LGG grade 3 | ||||||
| Mobadersany et al. (2018) [60] | Brain cancer | TCGA-GBM & LGG/1,061 (samples) | Survival analysis | CNN | Rotation, normalization | C-index 0.754 |
| Wu et al. (2018) [61] | Ovarian cancer | Biopsy/7,392 (images) | Classification Subtypes | CNN | Rotation, image enhancement | Overall classification acc. 78.2% |
| Zhang et al. (2017) [62] | Cervix cancer | HEMLBC/1,978 Herlev/917 (images) | Decision Benign/cancer | CNN | Rotation, translation, | Image level decision AUC 0.99 |
| Xu et al. (2017) [63] | Sickle cell disease | Red-blood cell/7,206 (patches) | Classification Cell types | CNN | Rotation, flip, translation, etc. | Cell level classification acc. 87.5% |
| Meier et al. (2018) [64] | Gastric cancer | TMA/469 (samples) CD8/Ki67 IHC | Survival analysis | CNN | None | Stratification by risk successful (p < .01) |
| Xie et al. (2016) [65] | - | Synthetic fluorescence microscopy cell/200 (images) | Cell counting | CNN | None | Mean absolute error < 2% |
| Tuominen et al. (2010) [66] | - | IHC stained breast cancer slides/100 | Cell counting | Comp. vision | None | Correlation coefficient 0.98 |
CNN, convolutional neural network; MIL, multiple instance learning; SVM, support vector machine; AUC, area under receiver operating characteristic curve; k-NN, k-nearest neighbor; WSI, whole slide image; CRF, Conditional random field; TCGA, The Cancer Genome Atlas; TMA, tissue microarray; IHC, immunohistochemistry; GBM, glioblastoma multiforme; LGG, lower grade glioma.

 E-submission
E-submission