Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 55(3); 2021 > Article
-
Original Article
Prognostic role of ALK-1 and h-TERT expression in glioblastoma multiforme: correlation with ALK gene alterations -
Dalia Elsers1, Doaa F. Temerik2, Alia M. Attia3, A. Hadia4, Marwa T. Hussien5

-
Journal of Pathology and Translational Medicine 2021;55(3):212-224.
DOI: https://doi.org/10.4132/jptm.2021.03.15
Published online: May 11, 2021
1Department of Pathology, Faculty of Medicine, Assiut University, Assiut, Egypt
2Department of Clinical Pathology, South Egypt Cancer Institute, Assiut University, Assiut, Egypt
3Department of Radiation Oncology, South Egypt Cancer Institute, Assiut University, Assiut, Egypt
4Department of Medical Oncology, South Egypt Cancer Institute, Assiut University, Assiut, Egypt
5Department of Oncologic Pathology, South Egypt Cancer Institute, Assiut University, Assiut, Egypt
- Corresponding Author: Marwa T. Hussien, MD, PhD, Department of Oncologic Pathology, South Egypt Cancer Institute, Assiut University, Assiut 171516, Egypt Tel: +20-88-208671, Fax: +20-088-2087709, E-mail: marwat.hussien@aun.edu.eg
© 20212020 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Figure & Data
References
Citations

- TERT Gene Mutation in Gliomas Cross‐Linked With (NTRK, PDL1, ALK, IDH, P53, EGFR, HER2): A Integrative Review TERT Gene Mutation in Gliomas
Gunter Gerson Santos, Guilherme Nobre Nogueira, Iasmin Maria Rodrigues Saldanha, Ana Gabriela Ponte Farias, Cauan Miranda Mateus, Osvaldo Mariano Viana Neto, Maria Jânia Teixeira
Journal of Surgical Oncology.2025; 131(6): 1202. CrossRef - Mapping chromatin remodelling in glioblastoma identifies epigenetic regulation of key molecular pathways and novel druggable targets
Claire Vinel, James Boot, Weiwei Jin, Nicola Pomella, Alexandra Hadaway, Charles Mein, Nicolae Radu Zabet, Silvia Marino
BMC Biology.2025;[Epub] CrossRef - Association of human telomerase reverse transcriptase promoter mutation with unfavorable prognosis in glioma: A systematic review and meta-analysis
Rongxuan Hua, Qiuxuan Li, Han Gao, Boya Wang, Chengwei He, Ying Wang, Sitian Zhang, Lei Gao, Hongwei Shang, Wen Wang, Jingdong Xu
Journal of Research in Medical Sciences.2023;[Epub] CrossRef - Immunohistochemical surrogates for molecular alterations for the classification and grading of gliomas
Viharkumar Patel, Sanda Alexandrescu
Seminars in Diagnostic Pathology.2022; 39(1): 78. CrossRef - Meme Kanseri Hastalarında hTERT Gen Ekspresyonunun Klinikopatolojik Önemi
Ebubekir DİRİCAN, Burak KANKAYA, Zeynep TATAR
Sağlık Bilimlerinde Değer.2022; 12(1): 22. CrossRef - Prognostic and predictive markers in glioblastoma and ALK overexpression
Jang-Hee Kim
Journal of Pathology and Translational Medicine.2021; 55(3): 236. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
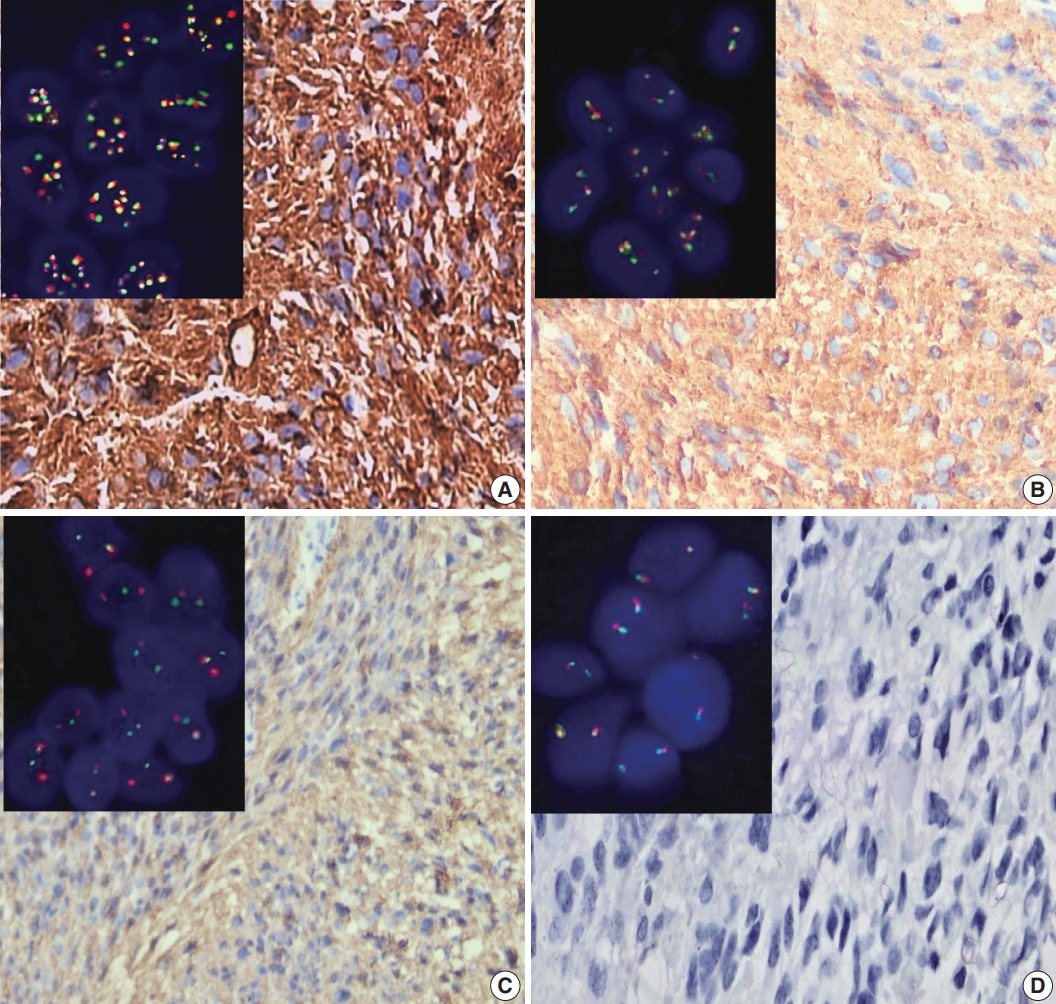
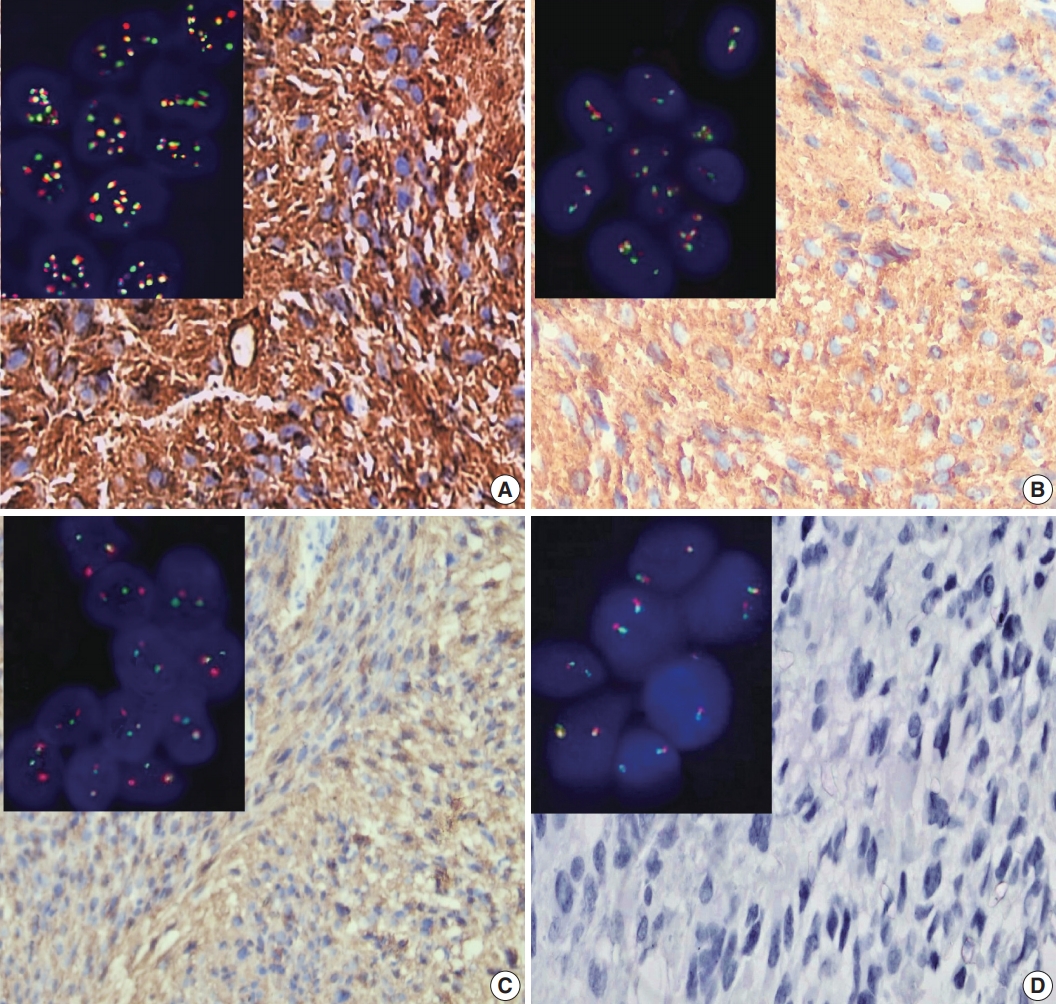
- Figure




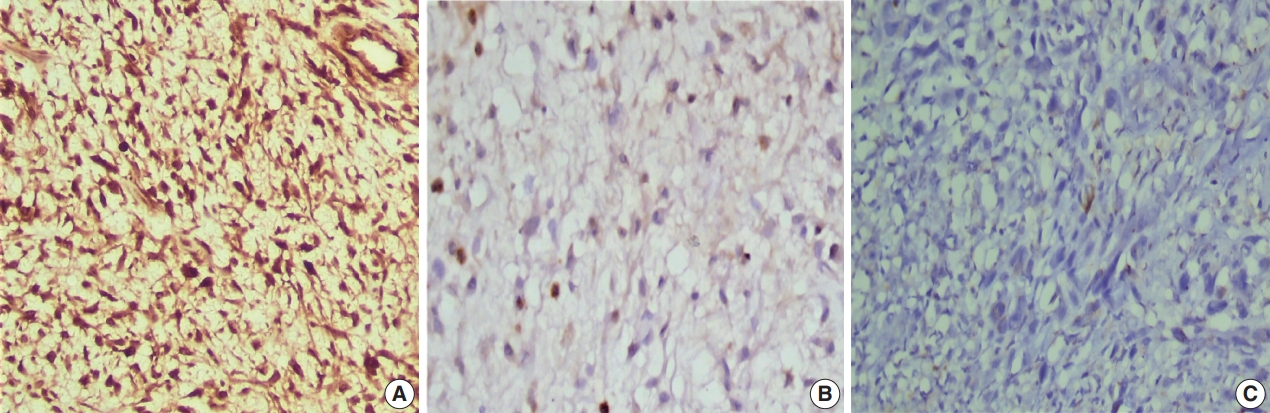
Fig. 1.
Fig. 2.
Fig. 3.
Fig. 4.
| Variable | No. (%) |
|---|---|
| Age (yr) | |
| < 50 | 17 (32.1) |
| ≥ 50 | 36 (67.9) |
| Sex | |
| Male | 36 (67.9) |
| Female | 17 (32.1) |
| Site | |
| CC | 4 (7.5) |
| FP | 12 (22.6) |
| PO | 15 (28.4) |
| TO | 7 (13.2) |
| PS | 6 (11.3) |
| TP | 9 (17.0) |
| Calcification | |
| Absent | 21 (39.6) |
| Present | 32 (60.4) |
| Multiplicity | |
| Single | 47 (88.7) |
| Multiple | 6 (11.3) |
| Tumor size | |
| Median (interquartile range) | 5 (4–7) |
| Type of surgical resection | |
| GTR | 19 (35.0) |
| STR | 25 (47.0) |
| Biopsy | 9 (17.0) |
| Ki LI (%) | |
| < 14 | 37 (69.8) |
| ≥ 14 | 16 (30.2) |
| Status | |
| Living | 21 (39.6) |
| Dead | 32 (60.4) |
| Therapeutic modalities | |
| RTH only | 12 (22.6) |
| CCRT | 14 (26.4) |
| CCRT and adjuvant TMZ | 27 (50.9) |
| Parameter | ALK-1 IHC |
h-TERT IHC |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Score 0 | Score 1 | Score 2 | Score 3 | p-value | Score 1 | Score 2 | Score 3 | p-value | |
| Age (yr) | .236 | .123 |
|||||||
| < 50 | 2 (11.8) | 6 (35.3) | 6 (35.3) | 3 (17.6) | 3 (17.6) | 8 (47.1) | 6 (35.3) | ||
| ≥ 50 | 6 (16.7) | 9 (25.0) | 7 (19.4) | 14 (38.9) | 5 (13.9) | 8 (22.2) | 23 (63.9) | ||
| Sex | .038 | .267 |
|||||||
| Male | 7 (19.4) | 6 (16.7) | 10 (27.8) | 13 (36.1) | 6 (16.7) | 13 (36.1) | 17 (47.2) | ||
| Female | 1 (5.9) | 9 (52.9) | 3 (17.6) | 4 (23.5) | 2 (11.8) | 3 (17.6) | 12 (70.6) | ||
| Site | .425 | .119 | |||||||
| CC | 2 (50.0) | 1 (25.0) | 0 | 1 (25.0) | 0 | 3 (75.0) | 1 (25.0) | ||
| FP | 1 (8.4) | 4 (33.3) | 4 (33.3) | 3 (25.0) | 2 (16.7) | 2 (16.7) | 8 (66.6) | ||
| PO | 2 (13.3) | 7 (46.7) | 2 (13.3) | 4 (26.7) | 5 (33.3) | 2 (13.3) | 8 (53.4) | ||
| TO | 2 (28.5) | 1 (14.3) | 3 (42.9) | 1 (14.3) | 1 (14.3) | 4 (57.1) | 2 (28.6) | ||
| PS | 0 | 0 | 2 (33.3) | 4 (66.7) | 0 | 1 (16.6) | 5 (83.4) | ||
| TP | 1 (11.2) | 2 (22.2) | 2 (22.2) | 4 (44.4) | 0 | 4 (44.4) | 5 (55.6) | ||
| Calcification | .315 | .016* | |||||||
| Absent | 1 (4.8) | 6 (28.6) | 5 (23.8) | 9 (42.8) | 3 (14.3) | 2 (9.5) | 16 (76.2) | ||
| Present | 7 (21.9) | 9 (28.1) | 8 (25.0) | 8 (25.0) | 5 (15.6) | 14 (43.8) | 13 (40.6) | ||
| Multiplicity | .046 | .501 | |||||||
| Single | 8 (17.1) | 14 (29.8) | 13 (27.7) | 12 (25.5) | 8 (17.0) | 15 (31.9) | 24 (51.1) | ||
| Multiple | 0 | 1 (16.7) | 0 | 5 (83.3) | 0 | 1 (16.7) | 5 (83.3) | ||
| Tumor size | .670 | .834 | |||||||
| <Median | 4 (14.2) | 6 (21.4) | 9 (32.2) | 9 (32.2) | 5 (17.8) | 7 (25.0) | 16 (57.2) | ||
| ≥ Median | 4 (14.2) | 9 (32.2) | 7 (25.0) | 8 (28.6) | 3 (12.0) | 9 (36.0) | 13 (52.0) | ||
| Type of surgical resection | .266 | .133 | |||||||
| GTR | 3 (15.8) | 4 (21.1) | 6 (31.6) | 6 (31.6) | 5 (26.3) | 3 (15.8) | 11 (57.9) | ||
| STR | 5 (20.0) | 7 (28.0) | 7 (28.0) | 6 (24.0) | 3 (12.0) | 11 (44.0) | 11 (44.0) | ||
| Biopsy | 0 | 4 (44.4) | 0 | 5 (55.6) | 0 | 2 (22.0) | 29 (54.7) | ||
| Ki LI (%) | < .001 | .005 | |||||||
| <14 | 8 (21.6) | 15 (40.5) | 10 (27.1) | 4 (10.8) | 8 (21.6) | 14 (37.8) | 15 (40.6) | ||
| ≥14 | 0 | 0 | 3 (18.7) | 13 (81.3) | 0 | 2 (12.5) | 14 (87.5) | ||
| Therapeutic modalities | .030 | .287 | |||||||
| RTH only | 0 | 3 (25.0) | 3 (25.0) | 6 (50.0) | 1 (8.3) | 2 (16.7) | 9 (75.0) | ||
| CCRT | 5 (35.7) | 4 (28.6) | 0 | 5 (35.7) | 4 (28.6) | 3 (21.4) | 7 (50.0) | ||
| CCRT and adjuvant TMZ | 3 (11.2) | 8 (29.6) | 10 (37.0) | 6 (22.2) | 3 (11.1) | 11 (40.7) | 13 (48.2) | ||
| ALK gene alterations |
p-value | ||||
|---|---|---|---|---|---|
| Negative | Rearrangement | Gain | Amplification | ||
| Age (yr) | .430 | ||||
| < 50 | 7 (41.2) | 7 (41.2) | 3 (17.6) | 0 | |
| ≥ 50 | 12 (33.3) | 11 (30.6) | 9 (25.0) | 4 (11.1) | |
| Sex | .181 | ||||
| Male | 10 (27.8) | 12 (33.3) | 10 (27.8) | 4 (11.1) | |
| Female | 9 (52.9) | 6 (35.3) | 2 (11.8) | 0 | |
| Site | .955 | ||||
| CC | 2 (50.0) | 1 (20.0) | 1 (20.0) | 0 | |
| FP | 4 (33.3) | 3 (25.0) | 3 (25.0) | 2 (16.7) | |
| PO | 6 (40.0) | 5 (33.3) | 3 (20.0) | 1 (6.7) | |
| TO | 2 (28.6) | 3 (42.8) | 1 (14.3) | 1 (14.3) | |
| PS | 2 (33.3) | 1 (16.7) | 3 (50.0) | 0 | |
| TP | 3 (33.3) | 5 (55.6) | 1 (11.1) | 0 | |
| Calcification | .710 | ||||
| Absent | 6 (28.6) | 7 (33.3) | 6 (28.6) | 2 (9.5) | |
| Present | 13 (40.6) | 11 (34.4) | 6 (18.8) | 2 (6.2) | |
| Multiplicity | .162 | ||||
| Single | 19 (40.4) | 15 (31.9) | 10 (21.3) | 3 (6.4) | |
| Multiple | 0 | 3 (50.0) | 2 (33.3) | 1 (16.7) | |
| Size | .836 | ||||
| < Median | 12 (38.7) | 9 (29.0) | 7 (22.6) | 3 (9.7) | |
| > Median | 7 (31.8) | 9 (40.9) | 5 (22.7) | 1 (4.6) | |
| Type of surgical resection | .125 | ||||
| GTR | 9 (47.4) | 3 (15.8) | 5 (26.3) | 2 (10.5) | |
| STR | 8 (32.0) | 9 (36.0) | 7 (28.0) | 1 (4.0) | |
| Biopsy | 2 (22.2) | 6 (66.7) | 0 | 1 (11.1) | |
| Ki- LI | .044 | ||||
| < 14% | 15 (40.5) | 15 (40.5) | 6 (16.2) | 1 (2.8) | |
| ≥ 14% | 4 (25.0) | 3 (18.8) | 6 (37.4) | 3 (18.8) | |
| ALK-1 IHC | .001 | ||||
| Score 0 | 8 (100) | 0 | 0 | 0 | |
| Score 1 | 6 (40.0) | 9 (60.0) | 0 | 0 | |
| Score 2 | 2 (15.4) | 5 (38.4) | 6 (46.2) | 0 | |
| Score 3 | 3 (17.7) | 4 (23.5) | 6 (35.3) | 4 (23.5) | |
| h-TERT IHC | .008 | ||||
| Score 1 | 6 (75.0) | 2 (25.0) | 0 | 0 | |
| Score 2 | 6 (37.5) | 9 (56.2) | 1 (6.3) | 0 | |
| Score 3 | 7 (24.1) | 7 (24.1) | 11 (38.0) | 4 (13.8) | |
| Therapeutic modalities | .027 | ||||
| RTH | 2 (16.7) | 3 (25.0) | 3 (25.0) | 4 (33.3) | |
| CCRT & adjuvant TMZ | 11 (40.7) | 10 (37.0) | 6 (22.3) | 0 | |
| CCRT | 6 (42.9) | 5 (35.7) | 3 (21.4) | 0 | |
| Spearman’s rho |
|||
|---|---|---|---|
| h-TERT expression | ALK-1 expression | ALK gene alterations | |
| h-TERT expression | |||
| Correlation coefficient | 1.000 | 0.602 | 0.476 |
| Sig. (2-tailed) | 0.002 |
0.007 |
|
| No. | 53 | 53 | 53 |
| ALK-1 expression | |||
| Correlation coefficient | 0.602 | 1.000 | 0.616 |
| Sig. (2-tailed) | 0.002 |
0.001 |
|
| No. | 53 | 53 | 53 |
| ALK gene alterations | |||
| Correlation coefficient | 0.476 | 0.616 | 1.000 |
| Sig. (2-tailed) | 0.007 |
0.001 |
|
| No. | 53 | 53 | 53 |
| Parameter | OS |
PFS |
||
|---|---|---|---|---|
| Log-rank (chi-square) | p-value | Log-rank (chi-square) | p-value | |
| Age (< 50 yr vs. ≥ 50 yr) | 3.146 | .076 | 2.419 | .120 |
| Sex (male vs. female) | 3.326 | .119 | 3.887 | .153 |
| Site (CC vs. FP vs. PO vs. TO vs. PS vs. TP) | 3.398 | .639 | 4.216 | .519 |
| Calcification (absent vs. present) | 0.666 | .414 | 0.253 | .615 |
| Multiplicity (single vs. multiple) | 3.993 | .254 | 3.505 | .190 |
| Size (<median vs. > median) | 0.010 | .921 | 0.048 | .827 |
| Type of surgical resection (GTR vs. STR vs. biopsy) | 0.066 | .967 | 0.168 | .919 |
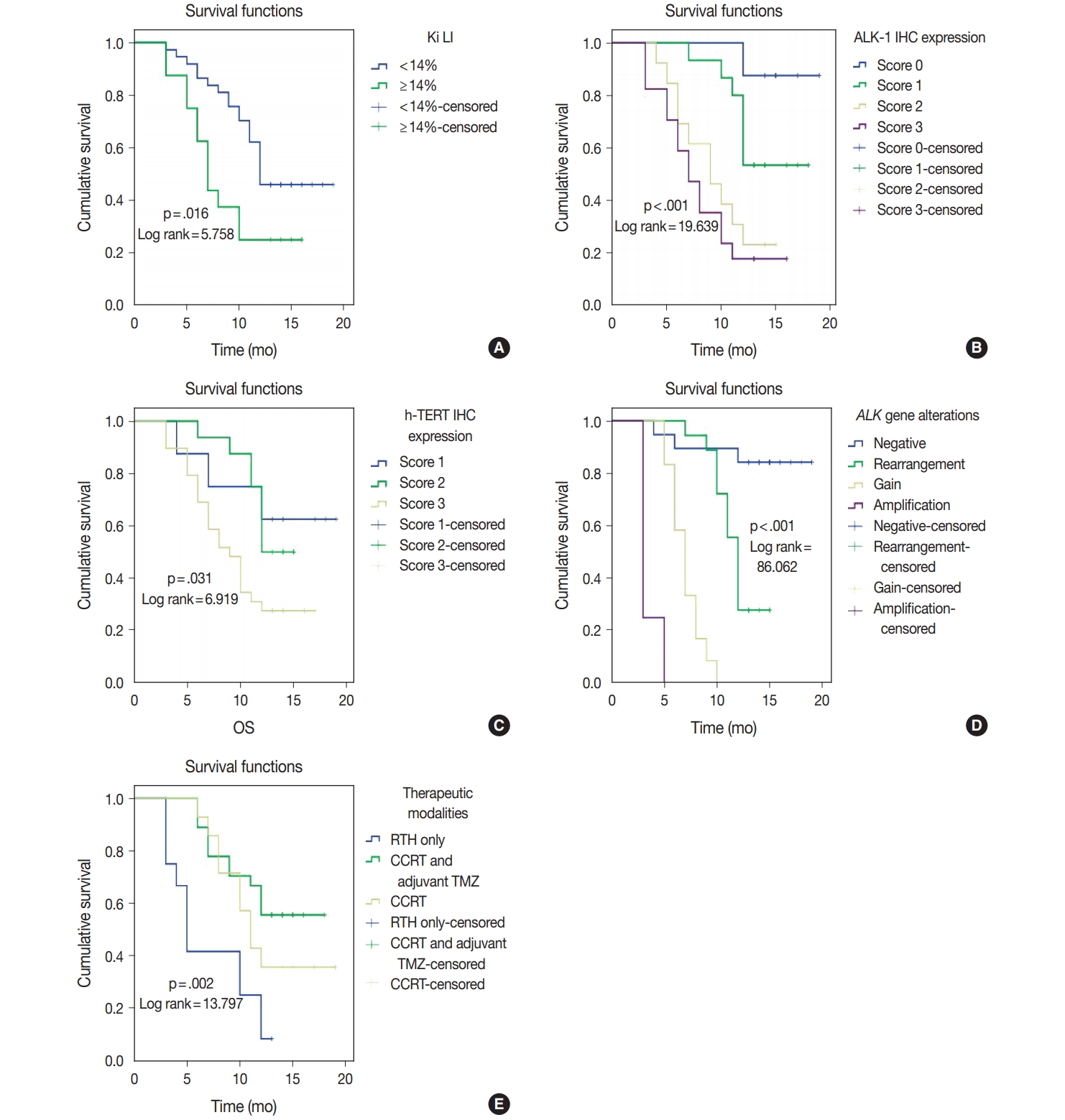
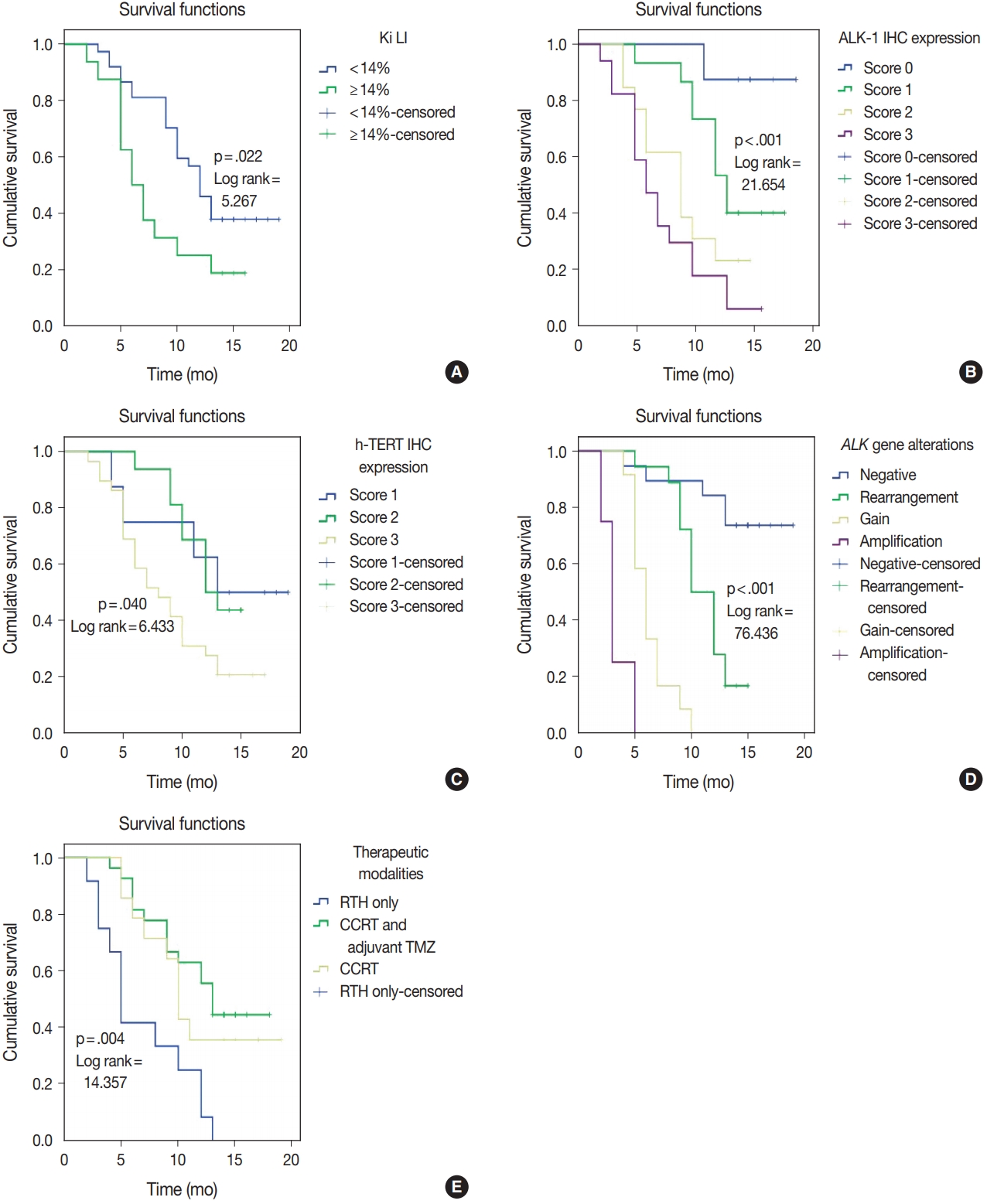
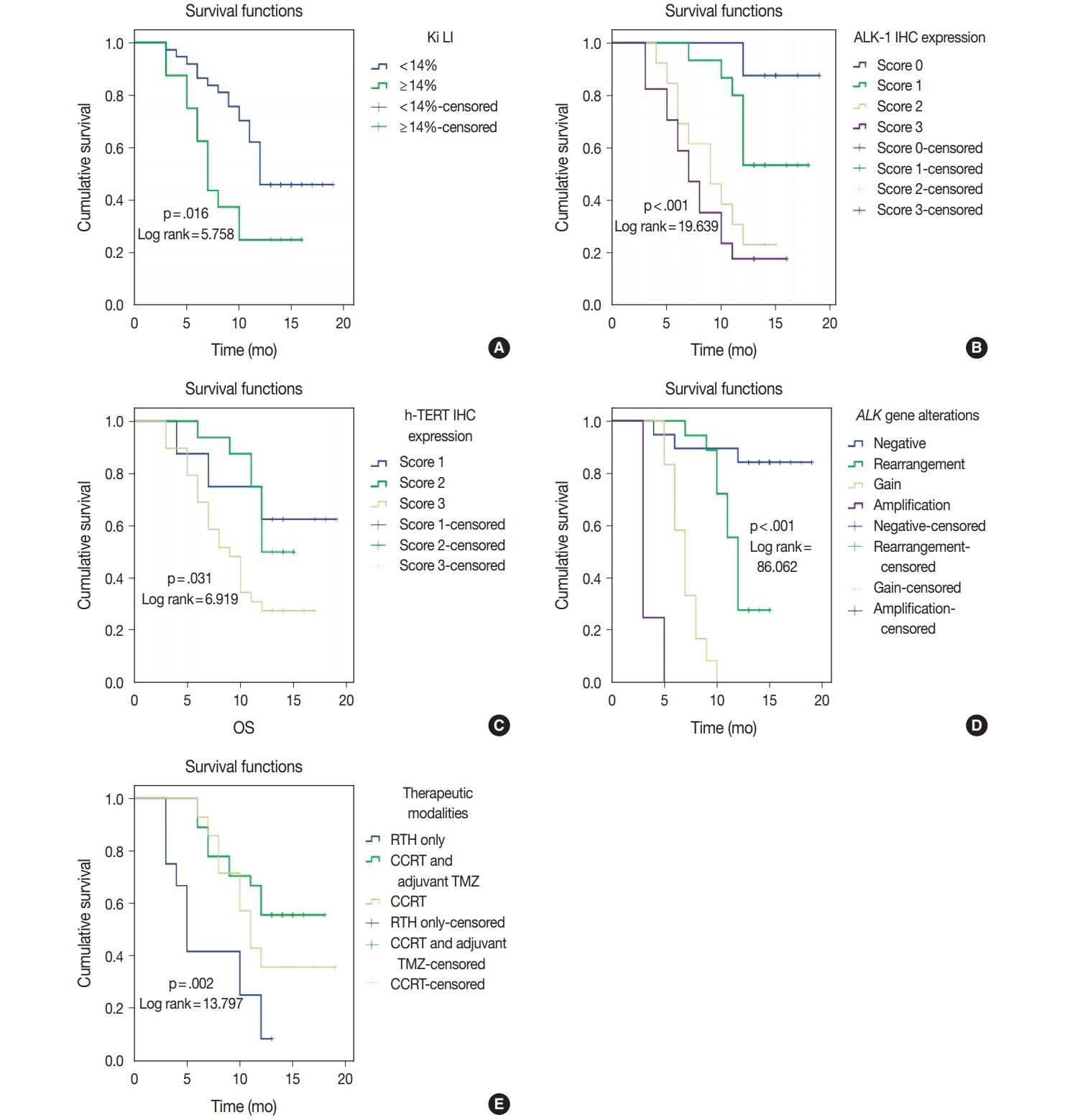
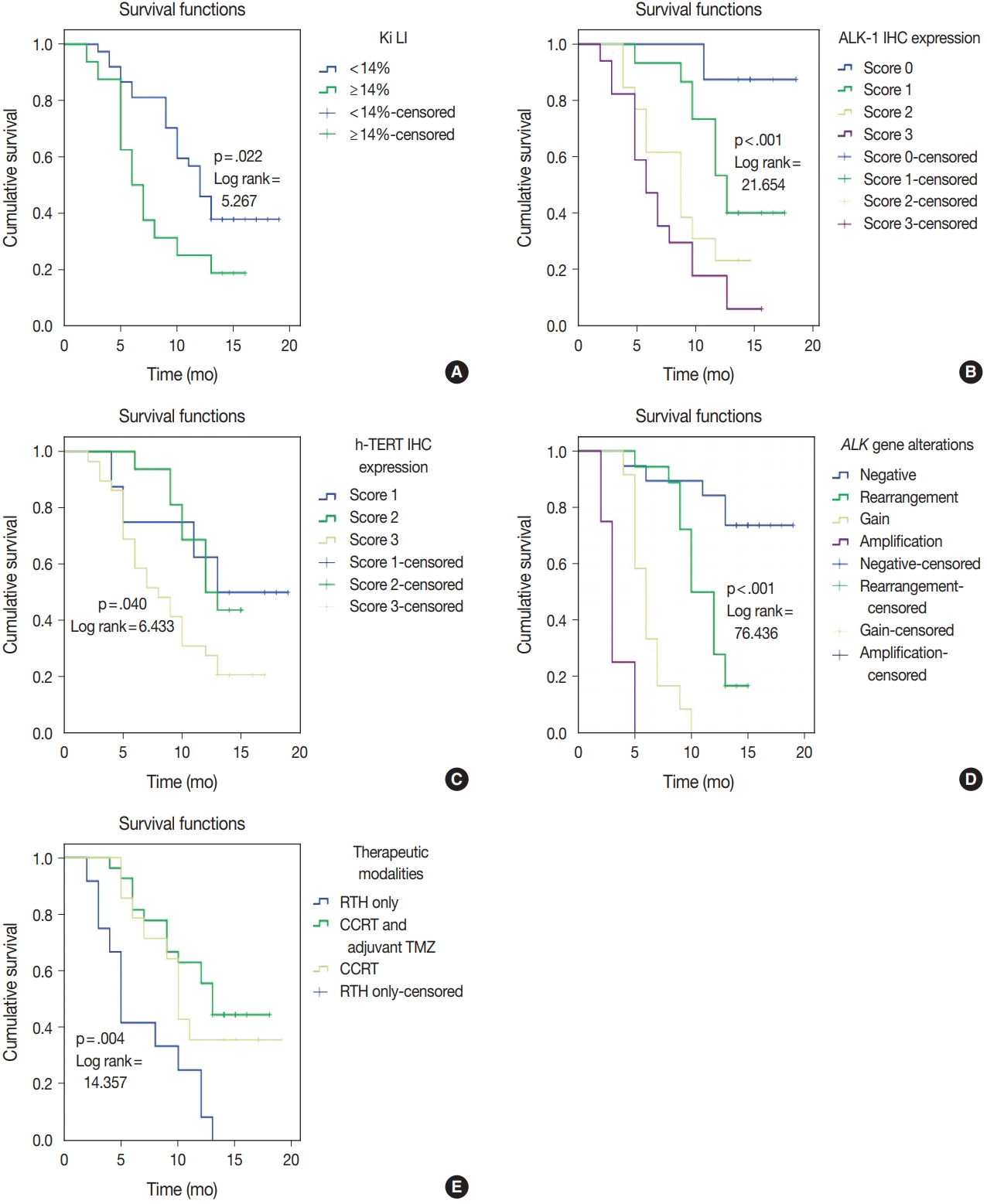
| Ki LI (< 14% vs. ≥ 14%) | 5.758 | .016 | 5.267 | .022 |
| ALK-1 IHC (score 0 vs. score 1 vs. score 2 vs. score 3) | 19.639 | < .001 | 21.654 | < .001 |
| h-TERT IHC (score 1 vs. score 2 vs. score 3) | 6.919 | .031 | 6.433 | .040 |
| ALK gene alterations (negative rearrangement vs. gain amplification) | 86.062 | < .001 | 76.436 | < .001 |
| Therapeutic modalities (RTH only vs. CCRT vs. CCRT and adjuvant TMZ) | 13.797 | .002 | 14.357 | .004 |
| OS |
PFS |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| B | SE | p-value | HR | 95 % CI for Exp (B) | B | SE | p-value | HR | 95 % CI for Exp (B) | |
| Ki LI | 0.125 | 0.468 | .790 | 1.133 | 0.453–2.833 | –0.001 | 0.435 | .998 | 0.099 | 0.426–2.344 |
| ALK-1 protein expression | 0.357 | 0.347 | .304 | 0.429 | 0.724–2.821 | 0.505 | 0.306 | .099 | 1.657 | 0.909–3.010 |
| h-TERT protein expression | –0.368 | 0.411 | .371 | 0.692 | 0.310–1.549 | –0.352 | 0.356 | .322 | 0.703 | 0.350–1.411 |
| ALK gene alterations | 2.017 | 0.421 | < .001 | 7.514 | 3.292–17.155 | 1.550 | 0.338 | < .001 | 4.711 | 2.429–9.136 |
| Therapeutic modalities | 0.050 | 0.311 | .872 | 1.052 | 0.571–1.935 | –0.007 | 0.295 | .982 | 0.993 | 0.558–1.769 |
CC, corpus callosum; FP, fronto-parietal; PO, parieto-occipital; TO, tempero-occipital; PS, parasagittal; TP, tempero-parietal; GTR, gross total resection; STR, subtotal resection; Ki LI, Ki labeling index; RTH, radiotherapy; CCRT, concurrent chemoradiotherapy; TMZ, temozolomide.
Significant at p < .05. ALK-1, anaplastic lymphoma kinase 1; h-TERT, human telomerase reverse transcriptase; IHC, immunohistochemistry; CC, corpus callosum; FP, fronto-parietal; PO, parieto-occipital; TO, tempero-occipital; PS, parasagittal; TP, tempero-parietal; GTR, gross total resection; STR, subtotal resection; Ki LI, Ki labeling index; RTH, radiotherapy; CCRT, concurrent chemoradiotherapy; TMZ, temozolomide. Fisher exact test was used in this table except when ≤20% of the cells have expected count less than 5 chi-square test was used instead.
Significant at p < .05. ALK-1, anaplastic lymphoma kinase 1; h-TERT, human telomerase reverse transcriptase; CC, corpus callosum; FP, fronto-parietal; PO, parieto-occipital; TO, tempero-occipital; PS, parasagittal; TP, tempero-parietal; GTR, gross total resection; STR, subtotal resection; Ki LI, Ki labeling index; IHC, immunohistochemistry; RTH, radiotherapy; CCRT, concurrent chemoradiotherapy; TMZ, temozolomide.
ALK-1, anaplastic lymphoma kinase 1; h-TERT, human telomerase reverse transcriptase; IHC, immunohistochemistry. Significant; correlation is significant at the 0.01 level (2-tailed).
Significant at p < 0.5. OS, overall survival; PFS, progression-free survival; CC, corpus callosum; FP, fronto-parietal; PO, parieto-occipital; TO, tempro-occipital; PS, parasagittal; TP, tempro-parietal; GTR, gross total resection; STR, subtotal resection; Ki LI, Ki labeling index; ALK-1, anaplastic lymphoma kinase 1; IHC, immunohistochemistry; h-TERT, human telomerase reverse transcriptase; RTH, radiotherapy; CCRT, concurrent chemoradiotherapy; TMZ, temozolomide.
Significant at p < 0.5. OS, overall survival; PFS, progression-free survival; HR, hazards ratio; CI, confident interval; Ki LI, Ki labeling index; ALK-1, anaplastic lymphoma kinase 1; h-TERT, human telomerase reverse transcriptase.

 E-submission
E-submission