Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 59(1); 2025 > Article
-
Original Article
Comparison of tissue-based and plasma-based testing for EGFR mutation in non–small cell lung cancer patients -
Yoon Kyung Kang1
 , Dong Hoon Shin1
, Dong Hoon Shin1 , Joon Young Park1
, Joon Young Park1 , Chung Su Hwang1
, Chung Su Hwang1 , Hyun Jung Lee1
, Hyun Jung Lee1 , Jung Hee Lee1
, Jung Hee Lee1 , Jee Yeon Kim1
, Jee Yeon Kim1 , JooYoung Na2
, JooYoung Na2
-
Journal of Pathology and Translational Medicine 2025;59(1):60-67.
DOI: https://doi.org/10.4132/jptm.2024.10.01
Published online: January 15, 2025
1Department of Pathology, Pusan National University School of Medicine, Yangsan, Korea
2Department of Forensic Medicine, Pusan National University School of Medicine, Yangsan, Korea
- Corresponding Author Dong Hoon Shin, MD, PhD Department of Pathology, Pusan National University School of Medicine, 49 Busandaehak-ro, Mulgeum-eup, Yangsan 50612, Korea Tel: +82-55-360-1859 Fax: +82-55-360-1855 E-mail: donghshin@pusan.ac.kr
© The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
- 5,085 Views
- 202 Download
Abstract
-
Background
- Epidermal growth factor receptor (EGFR) gene mutation testing is crucial for the administration of tyrosine kinase inhibitors to treat non–small cell lung cancer. In addition to traditional tissue-based tests, liquid biopsies using plasma are increasingly utilized, particularly for detecting T790M mutations. This study compared tissue- and plasma-based EGFR testing methods.
-
Methods
- A total of 248 patients were tested for EGFR mutations using tissue and plasma samples from 2018 to 2023 at Pusan National University Yangsan Hospital. Tissue tests were performed using PANAmutyper, and plasma tests were performed using the Cobas EGFR Mutation Test v2.
-
Results
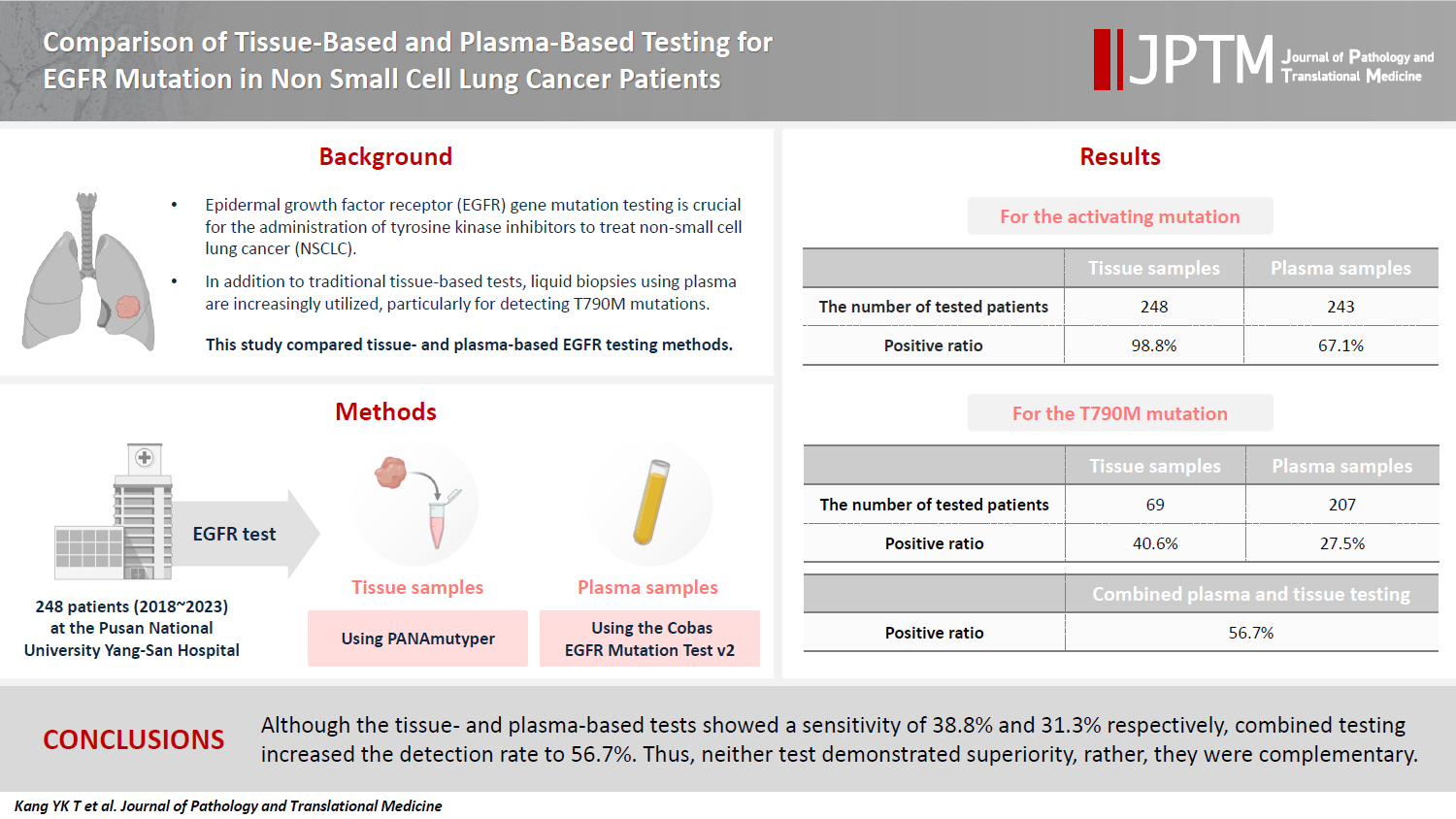
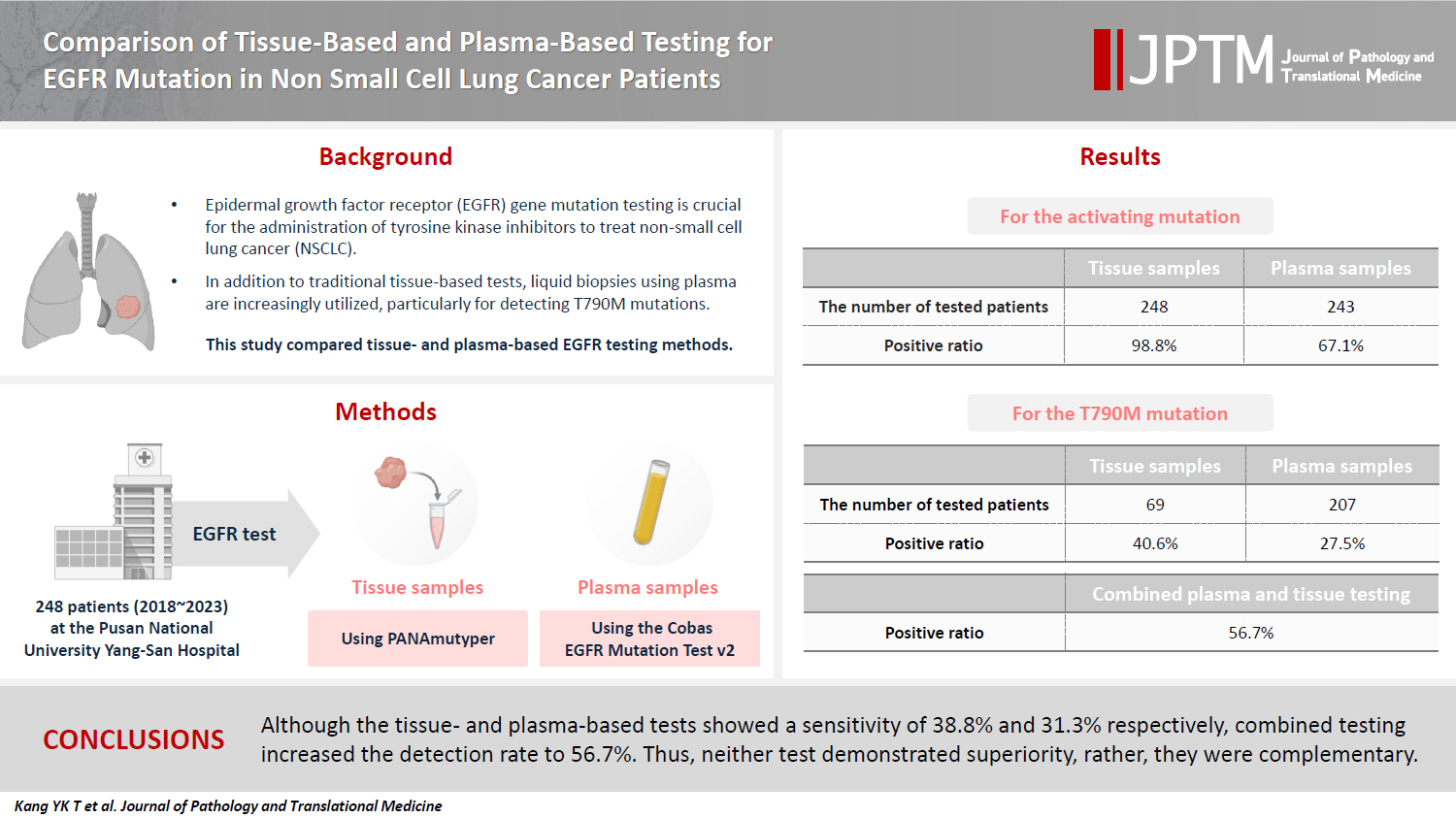
- All 248 patients underwent tissue-based EGFR testing, and 245 (98.8%) showed positive results. Of the 408 plasma tests, 237 (58.1%) were positive. For the T790M mutation, tissue biopsies were performed 87 times in 69 patients, and 30 positive cases (38.6%) were detected. Plasma testing for the T790M mutation was conducted 333 times in 207 patients, yielding 62 positive results (18.6%). Of these, 57 (27.5%) were confirmed to have the mutation via plasma testing. Combined tissue and plasma tests for the T790M mutation were positive in nine patients (13.4%), while 17 (25.4%) were positive in tissue only and 12 (17.9%) in plasma only. This mutation was not detected in 28 patients (43.3%).
-
Conclusions
- Although the tissue- and plasma-based tests showed a sensitivity of 37.3% and 32.8%, respectively, combined testing increased the detection rate to 56.7%. Thus, neither test demonstrated superiority, rather, they were complementary.
- Non–small cell lung cancer (NSCLC) is a major cause of cancer-related mortality globally [1,2]. However, the patients' prognosis and quality of life have significantly improved with the introduction of targeted therapies, including tyrosine kinase inhibitors (TKIs), which act against mutations in the epidermal growth factor receptor (EGFR). Despite their initial effectiveness, resistance to first- or second-generation TKIs often develops within months or years owing to various mechanisms, with the T790M mutation in the EGFR gene accounting for approximately 50% of these resistance cases [3,4]. To counter this, third-generation TKIs such as osimertinib have been developed to target this specific mutation, making detection of the T790M mutation crucial for patients with NSCLC who have developed resistance to previous TKIs [3-9]. However, this detection can be challenging, especially in patients with small, hard-to-reach tumors, or when tissue biopsy is not an option.
- Recent developments have underscored the potential of liquid biopsy as either an alternative or a supplementary approach to tissue biopsy for molecular diagnostics, including the detection of the T790M mutation in blood samples [5-9]. This method is particularly appealing because of its noninvasive nature, feasibility of repeated testing, fast turnaround time, and potential to reflect changes in tumor burden through serial monitoring. Unlike tissue biopsies, which only reveal mutations at the sampling site, blood tests can detect all tumor DNA present in patient bloodstreams. Currently, the National Comprehensive Cancer Network guidelines recommend liquid biopsy as an alternative to tissue biopsy for initial T790M mutation testing [10]. The importance of accurately determining the sensitivity and specificity of liquid biopsies is paramount, given their broad applications. However, optimization of liquid biopsies is challenging, primarily because of the inability to directly quantify cell-free tumor DNA (ctDNA). This uncertainty in ctDNA levels complicates quality control compared with that of tissue biopsies [8,9]. In contrast, tissue biopsies allow pathologists to directly observe the number of tumor cells under a microscope, providing a clearer understanding of whether negative results stem from an insufficient amount of tumor DNA. Direct observation is not possible using liquid biopsies. Therefore, these differences may play a role in the diverse findings regarding the effectiveness of liquid biopsy for EGFR mutation detection, as reported in various studies [3-9].
- In Korea, tissue-based tests are performed in the Department of Pathology, whereas blood-based tests are performed either in the Department of Pathology or the Department of Laboratory Medicine, depending on the institution [8,9,11]. However, this division may result in communication challenges between the two departments, potentially affecting the management of EGFR testing. The primary objective of our study was to evaluate the performance of tests performed independently in each department. Furthermore, we evaluated the effectiveness of EGFR assays conducted on both tissue and blood samples by examining a series of consecutive tests from each patient. This approach enabled us to ascertain the overall sensitivity and specificity on an individual basis, rather than limiting our evaluation to a per-test perspective. Consequently, our study also aimed to offer comprehensive insights into the detection of T790M mutations in both tissue and liquid biopsies. This was aimed at assisting clinicians in choosing the most appropriate testing approach for patients considering variables such as tumor burden and metastatic sites.
INTRODUCTION
- Patients
- Between 2018 and 2023, a total of 590 plasma EGFR mutation tests were conducted at Pusan National University Yangsan Hospital. Among these patients, those diagnosed with adenocarcinoma, squamous cell carcinoma, or NSCLC were selected. Subsequently, only patients who also underwent an EGFR mutation test using tissue-based methods and tested positive for an EGFR mutation in either tissue or plasma at least once were included in the study. For analysis of T790M mutation, only the patient who received TKI treatment were considered. A comprehensive review of the longitudinal data of these patients was performed, including pathology reports with EGFR tests, the results of plasma EGFR tests, radiological imaging, and positron emission tomography scans.
- Tissue EGFR test
- The EGFR test in tissue was performed using the PANAMuytper, a peptide nucleic acid (PNA)–mediated, real-time polymerase chain reaction (PCR)–based assay. Mutant DNA was selectively amplified using wild-type DNA-specific PNA clamp probes, after which mutant-specific PNA detection probes were used to genotype the EGFR mutations using fluorescence melting curve analysis. This assay can detect and discriminate 47 types of EGFR mutations, including three G719X substitutions, 29 exon 19 deletions (Exon19del), one T790M substitution, one S768I substitution, 10 exon 20 insertions (Exon20ins), two L858R substitutions, and one L861Q substitution, with a high level of sensitivity [12].
- Plasma EGFR test
- Venous blood samples were collected from patients using 21G needles in a 10-mL Cell-Free DNA BCT tube (Streck, Omaha, NE, USA) per patient. For the 1-step centrifugation group, within 4 hours of room-temperature blood collection, blood samples were centrifuged at 1,600 ×g for 10 minutes, and 2 mL was dispensed into each Eppendorf tube. EGFR mutations were identified using the Cobas EGFR Mutation Test v2 on a Cobas z 480 analyzer (Roche Diagnostics, Pleasanton, CA, USA), according to the manufacturer’s instructions [11]. This assay was designed to detect insertions and deletions in EGFR, such as deletions in exon 19 and insertions in exon 20, as well as nucleic acid substitutions, such as G719X, S768I, T790M, L858R, and L861Q.
- Statistical analyses
- Python software ver. 3.7.6 was used to perform all computation and analyses in this study. The Pandas library was used to perform basic statistical analysis of the data. All statistical tests were two-sided, and p < .05 was considered significant unless otherwise specified.
MATERIALS AND METHODS
- Clinical characteristics of patients
- A total of 248 patients were enrolled for this analysis (106 males and 142 females) (Table 1). The average age at diagnosis was 67 years (range, 40 to 86 years). The pathological diagnoses included 238 cases of adenocarcinoma, eight of squamous cell carcinoma, and two of adenosquamous carcinoma. The distribution of patients according to stage was as follows: stage I, 16; stage II, 10; stage III, 39; and stage IV, 183. The most common sites of metastasis upon recurrence or progression were the lungs (136 patients), followed by the brain (n = 74), bone (n = 52), pleura (n = 31), lymph nodes (n = 19), liver (n = 15), and other organs (n = 10).
- Activating EGFR mutation
- All 248 enrolled patients underwent EGFR testing using tissue samples, including biopsy or resection, at the time of diagnosis or during the follow-up period. The results identified 245 (98.8%) positive and three (1.2%) negative cases (Table 2). For plasma EGFR testing, 408 tests were conducted across these patients, with 58 at the time of diagnosis and 350 following the initial diagnosis, showing 237 positive results (58.1%) and 171 negative results (41.9%). Of the patients tested, 163 (67.1%) tested positive and 80 (32.6%) tested negative in the plasma tests (Table 2). When both tissue- and plasma-based tests yielded positive results, they were consistent with exon 19 deletion, L858R, exon 20 insertion, and L861Q mutations. However, discrepancies were observed in compound mutations. In these instances, tissue tests identifying exon 19 deletion/L861Q, L861Q/G719S, G719S/S768, and L858R/T790M mutations were detected as exon 19 deletion, L861Q, G719S/L861Q, and L858R mutations, respectively, in the plasma-based tests. The tissue-based tests did not detect activating mutations in four of the samples. In one sample, the tumor cell count ranged from 200 to 500, while the other three samples had tumor cell counts exceeding 1,000. A repeated tissue biopsy conducted after 18 months in one case revealed an exon 19 deletion mutation. Additionally, three other cases were found to have mutations in plasma-based tests: one with an exon 19 deletion and the others with the L858R mutation.
- T790M mutation
- Of the 248 enrolled patients, 210 received TKI treatment. Among these, 69 underwent a tissue biopsy, 207 had a plasma test to detect the T790M mutation, and 66 underwent both tests. Tissue biopsies were performed 87 times in 69 patients to detect the T790M mutation, which resulted in 30 (34.1%) positive results. Of the 69 patients, 28 (40.6%) harbored the T790M mutation. Plasma tests for the T790M mutation were conducted 333 times in 207 patients, with 62 positive results (18.6%). Of 271 negative test results, activating EGFR mutations were not detected in 148 patients. Among the 207 patients, 57 (27.5%) were confirmed to harbor the T790M mutation by plasma testing. Of these, 42 (20.3%) tested positive in the first plasma test, 10 out of 83 patients (12%) tested positive in the second test, and five out of 33 patients (15.2%) tested positive in the third test. Regarding tissue biopsies, 53 patients underwent the procedure once, 14 patients underwent it twice, and two patients underwent it thrice. For plasma tests during the treatment course, 124 patients were tested once, 50 patients twice, 24 patients thrice, eight patients four times, and one patient five times.
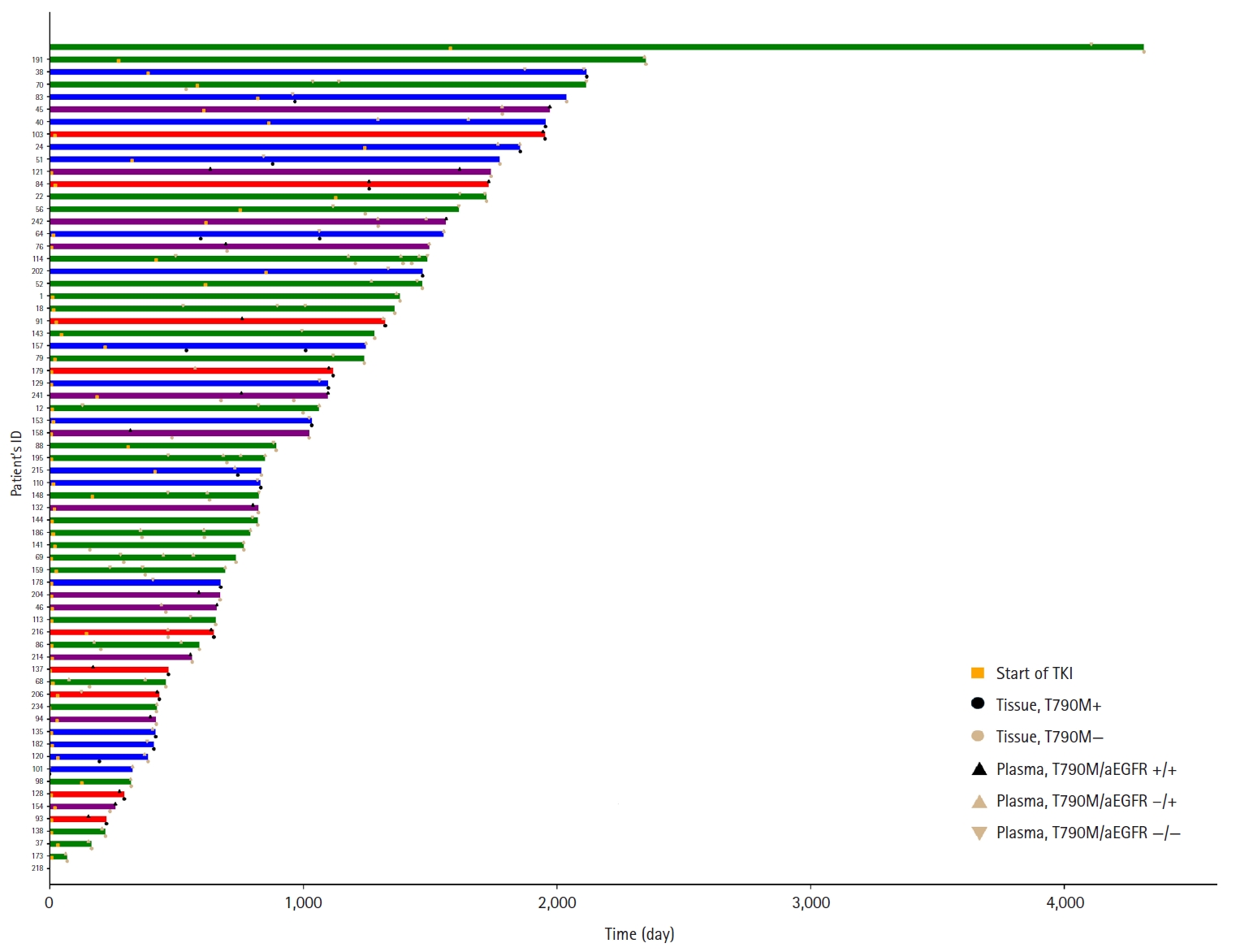
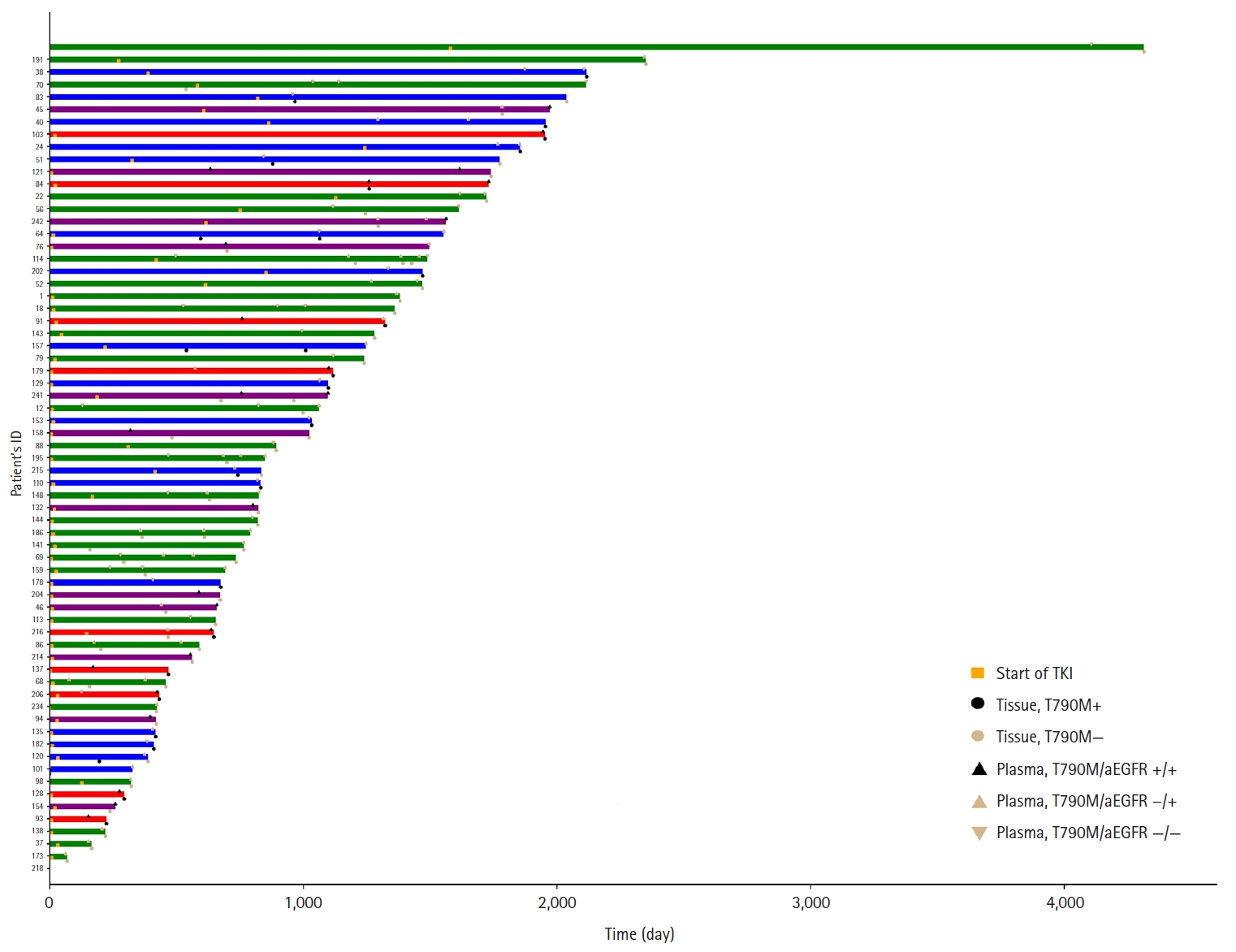
- Sixty-six patients underwent both tissue- and plasma-based EGFR testing for T790M mutation (Table 3, Fig. 1). In nine (13.4%) of these patients, the T790M mutation was detected in both tests. The mutation was identified exclusively in the tissue samples of 17 patients (25.4%) and in the plasma samples of 12 patients (17.9%). In 28 patients (43.3%), neither test detected the T790M mutation. These cases are illustrated in Fig. 1.
- Correlation of EGFR mutation detection with clinical parameters
- In patients who underwent plasma EGFR testing at the time of histological diagnosis, a positive plasma test result for activating mutations was associated with the presence of nodal or distant metastases (Table 4). Conversely, the detection of the T790M mutation in the plasma did not exhibit a correlation with either nodal status or distant metastasis (Table 4).
RESULTS
- Liquid biopsy is an emerging and highly valuable tool for detecting actionable mutations. Numerous studies have demonstrated its utility and yielded promising results [13-17]. One of the most widely used liquid biopsy methods is the plasma-based EGFR test for patients with lung cancer, particularly to guide TKI treatment decisions. Therefore, evaluating the efficacy of this test is of paramount importance as it directly influences treatment choices. Although several studies have demonstrated that the plasma-based EGFR test is comparable to the tissue-based test, the reported detection rates of the T790M mutation in these studies vary, ranging from 19.2% to 47.1% [13-16]. In this comprehensive study, data from 248 patients who underwent both tissue- and plasma-based EGFR tests to detect EGFR mutations were analyzed. To identify T790M mutations, 333 plasma tests were performed on 207 patients treated with TKI, yielding positive results in 62 tests (18.6%) and confirming the T790M mutation in 57 patients (27.4%). Conversely, tissue-based tests were performed 87 times in 69 patients, resulting in 30 positive tests (34.1%), indicating the presence of the T790M mutation in 28 patients (38.6%). In 66 patients subjected to both tissue- and plasma-based EGFR tests, tissue-based tests demonstrated a marginally higher sensitivity, yielding positive results in 26 patients (38.8%) compared with 21 patients (31.3%) for plasma-based tests. Nevertheless, the sensitivity of both methods was relatively low considering the estimated prevalence of the T790M mutation in 50% of TKI resistance cases [3-6]. Interestingly, when the results from both tests were combined, the positive rate increased to 38 patients (56.7%), exceeding the expected prevalence of 50%. These findings suggest that tissue- and plasma-based tests complement each other.
- In our study, 408 plasma assays were conducted to detect EGFR-activating mutations in a cohort of 248 patients with confirmed mutations. Of these, only 237 (58.1%) were positive. Remarkably, 85 of the 248 patients tested negative for these mutations in the plasma, despite the ability of plasma testing to facilitate repeated measurements. Furthermore, an examination of the 272 assays that were negative for the T790M mutation indicated that 148 (54.4%) failed to detect the activating mutation, suggesting that over half of the tests that failed to detect the activating EGFR mutation may not reliably indicate a true-negative result for the T790M mutation. Considering that there were no instances in which the T790M mutation was detected without the concurrent detection of the activating mutation, the presence of the activating mutation was a reliable indicator of ctDNA levels. Our findings suggest that ctDNA levels frequently fall below the detection threshold of the assay, indicating that liquid biopsies for other genes may also face similar challenges and thus require high sensitivity for effective clinical application.
- The prevalent understanding is that significant tumor mass is crucial for the abundant presence of ctDNA in the bloodstream. Our study showed that higher N and M statuses at initial diagnosis were associated with the detection of activating EGFR mutations in the plasma, with a notably stronger correlation observed for M status. This relationship continues even after the initiation of TKI therapy, where the original M status significantly predicts the presence of activating mutations, suggesting that an increased tumor burden may lead to higher ctDNA levels. However, no significant link was found between T790M mutation detection and T, N, and M statuses. To explore the factors influencing the detection of the T790M mutation, we analyzed the organs that were predominantly affected as the cancer progressed. Our data showed that the lungs were the most affected organs, followed by the brain, bones, pleura, lymph nodes, liver, and other organs. Moreover, there was no association between the presence of the T790M mutation and specific organs affected by metastasis or the overall number of involved organs. While TNM status and the number of affected organs might reflect the patient tumor load, which is expected to correlate with ctDNA levels, the absence of a connection with the T790M mutation indicates a potential need for more sensitive detection techniques, such as digital droplet PCR or next-generation sequencing (NGS), or that factors other than tumor load might affect the detectability of the mutation in plasma. Regarding tissue-based T790M testing, among the 30 positive cases, 22 originated from the lungs or bronchi. In contrast, of the 60 negative cases, 33 samples were collected from these locations, suggesting that the biopsy site did not significantly affect the test results.
- The primary advantage of plasma-based testing is its noninvasive nature, which allows for repeated testing and increases the likelihood of a positive result. In our investigation, initial plasma screening revealed the T790M mutation in 42 of 207 patients, yielding a positivity rate of 20.2%. A subsequent plasma test of 83 patients identified 10 more individuals (12.0%) harboring this mutation. In the third round of testing involving 33 patients, the mutation was found in five individuals, resulting in a positivity rate of 15.2%. In contrast, the first round of tissue-based testing demonstrated a considerably higher positivity rate, with 25 of the 69 patients (35.7%) testing positive for the T790M mutation. However, a follow-up tissue-based assessment of 18 patients showed a reduced positivity rate of 11.1%, with just two patients testing positive. This trend suggests a decrease in positivity rates for both plasma- and tissue-based tests in subsequent rounds compared with initial testing. Owing to the noninvasive nature of plasma-based testing, it is recommended to start with this method for detecting the T790M mutation. If the initial plasma test result is negative, a tissue-based test should be considered owing to its higher success rate. In contrast to the ease of plasma testing, tissue biopsies offer additional advantages, including the ability to identify resistance mechanisms beyond the T790M mutation, such as histological transformation. The transition to small cell carcinoma is a well-established example of such a transformation.
- Although a limitation of this study was that tissue- and plasma-based tests were performed on different platforms, previous studies have shown that both platforms have similar overall performance in terms of EGFR mutation detection [18]. In conclusion, this study suggests that both methods are complementary for detecting the T790M mutation, as neither method was completely satisfactory. Plasma-based tests frequently fail to detect activating mutations; therefore, clinicians should be aware of this drawback and perform tissue biopsy in such cases. Currently, the selection of test modalities, timing, and biopsy sites depends solely on the judgment of the clinician, with a lack of available tools to predict the outcome of these decisions. Recent advancements in highly sensitive techniques, including NGS and digital droplet PCR, have the potential to enhance the success rates of detecting T790M mutations in both tissue- and plasma-based tests.
DISCUSSION
Ethics Statement
This study was approved by the Institutional Review Board (IRB) of Pusan National University Yangsan Hospital (No. 2024-0021). The requirement for informed consent was waived by the IRB.
Availability of Data and Material
All data generated or analyzed during the study are included in this published article.
Code Availability
Not applicable.
Author Contributions
Conceptualization: DHS. Data curation: YKK. Formal analysis: YKK, JYP, CSH. Funding acquisition: DHS. Supervision: DHS. Writing—original draft: YKK, HJL. Writing—review & editing: JHL, JYK, JN, DHS. Approval of the final manuscript: all authors.
Conflicts of Interest
The authors declare that they have no potential conflicts of interest.
Funding Statement
This work was supported by a 2-year research grant of Pusan National University.
- 1. Wang M, Herbst RS, Boshoff C. Toward personalized treatment approaches for non-small-cell lung cancer. Nat Med 2021; 27: 1345-56. ArticlePubMedPDF
- 2. Howlader N, Forjaz G, Mooradian MJ, et al. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med 2020; 383: 640-9. ArticlePubMedPMC
- 3. Wu YL, Tsuboi M, He J, et al. Osimertinib in resected EGFR-mutated non-small-cell lung cancer. N Engl J Med 2020; 383: 1711-23. ArticlePubMed
- 4. Vaclova T, Grazini U, Ward L, et al. Clinical impact of subclonal EGFR T790M mutations in advanced-stage EGFR-mutant non-small-cell lung cancers. Nat Commun 2021; 12: 1780.ArticlePubMedPMCPDF
- 5. Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA analysis for detection of the EGFR T790M mutation in patients with advanced non-small cell lung cancer. J Thorac Oncol 2017; 12: 1061-70. ArticlePubMed
- 6. Naka G, Yokoyama T, Usui K, et al. Final report on plasma ctDNA T790M monitoring during EGFR-TKI treatment in patients with EGFR mutant non-small cell lung cancer (JP-CLEAR trial). Jpn J Clin Oncol 2022; 52: 791-4. ArticlePubMedPMCPDF
- 7. Rolfo C, Mack P, Scagliotti GV, et al. Liquid biopsy for advanced NSCLC: a consensus statement from the International Association for the Study of Lung Cancer. J Thorac Oncol 2021; 16: 1647-62. ArticlePubMed
- 8. Shin DH, Shim HS, Kim TJ, et al. Provisional guideline recommendation for EGFR gene mutation testing in liquid samples of lung cancer patients: a proposal by the Korean Cardiopulmonary Pathology Study Group. J Pathol Transl Med 2019; 53: 153-8. ArticlePubMedPMCPDF
- 9. Chang S, Hur JY, Choi YL, Lee CH, Kim WS. Current status and future perspectives of liquid biopsy in non-small cell lung cancer. J Pathol Transl Med 2020; 54: 204-12. ArticlePubMedPMCPDF
- 10. Ettinger DS, Wood DE, Aisner DL, et al. Non-small cell lung cancer, version 3.2022, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2022; 20: 497-530. ArticlePubMed
- 11. Shin KH, Lee SM, Park K, et al. Effects of different centrifugation protocols on the detection of EGFR mutations in plasma cell-free DNA. Am J Clin Pathol 2022; 158: 206-11. ArticlePubMedPMCPDF
- 12. Lee J, Han YB, Kwon HJ, Lee SK, Kim H, Chung JH. Landscape of EGFR mutations in lung adenocarcinoma: a single institute experience with comparison of PANAMutyper testing and targeted next-generation sequencing. J Pathol Transl Med 2022; 56: 249-59. ArticlePubMedPMCPDF
- 13. Jung HA, Ku BM, Kim YJ, et al. Longitudinal monitoring of circulating tumor DNA from plasma in patients with curative resected stages I to IIIA EGFR-mutant non-small cell lung cancer. J Thorac Oncol 2023; 18: 1199-208. ArticlePubMed
- 14. Karlovich C, Goldman JW, Sun JM, et al. Assessment of EGFR mutation status in matched plasma and tumor tissue of NSCLC patients from a phase I study of rociletinib (CO-1686). Clin Cancer Res 2016; 22: 2386-95. ArticlePubMedPMCPDF
- 15. Mack PC, Miao J, Redman MW, et al. Circulating tumor DNA kinetics predict progression-free and overall survival in EGFR TKI-treated patients with EGFR-mutant NSCLC (SWOG S1403). Clin Cancer Res 2022; 28: 3752-60. ArticlePubMedPMCPDF
- 16. Kok PS, Lee K, Lord S, et al. Clinical utility of plasma EGFR mutation detection with quantitative PCR in advanced lung cancer: a meta-analysis. Lung Cancer 2021; 154: 113-7. ArticlePubMed
- 17. Minari R, Mazzaschi G, Bordi P, et al. Detection of EGFR-activating and T790M mutations using liquid biopsy in patients with EGFR-mutated non-small-cell lung cancer whose disease has progressed during treatment with first- and second-generation tyrosine kinase inhibitors: a multicenter real-life retrospective study. Clin Lung Cancer 2020; 21: e464-73. ArticlePubMed
- 18. Han AL, Kim HR, Choi KH, et al. Comparison of cobas EGFR Mutation Test v2 and PANAMutyper-R-EGFR for detection and semi-quantification of epidermal growth factor receptor mutations in plasma and pleural effusion supernatant. Ann Lab Med 2019; 39: 478-87. ArticlePubMedPMCPDF
REFERENCES
Figure & Data
References
Citations

 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure
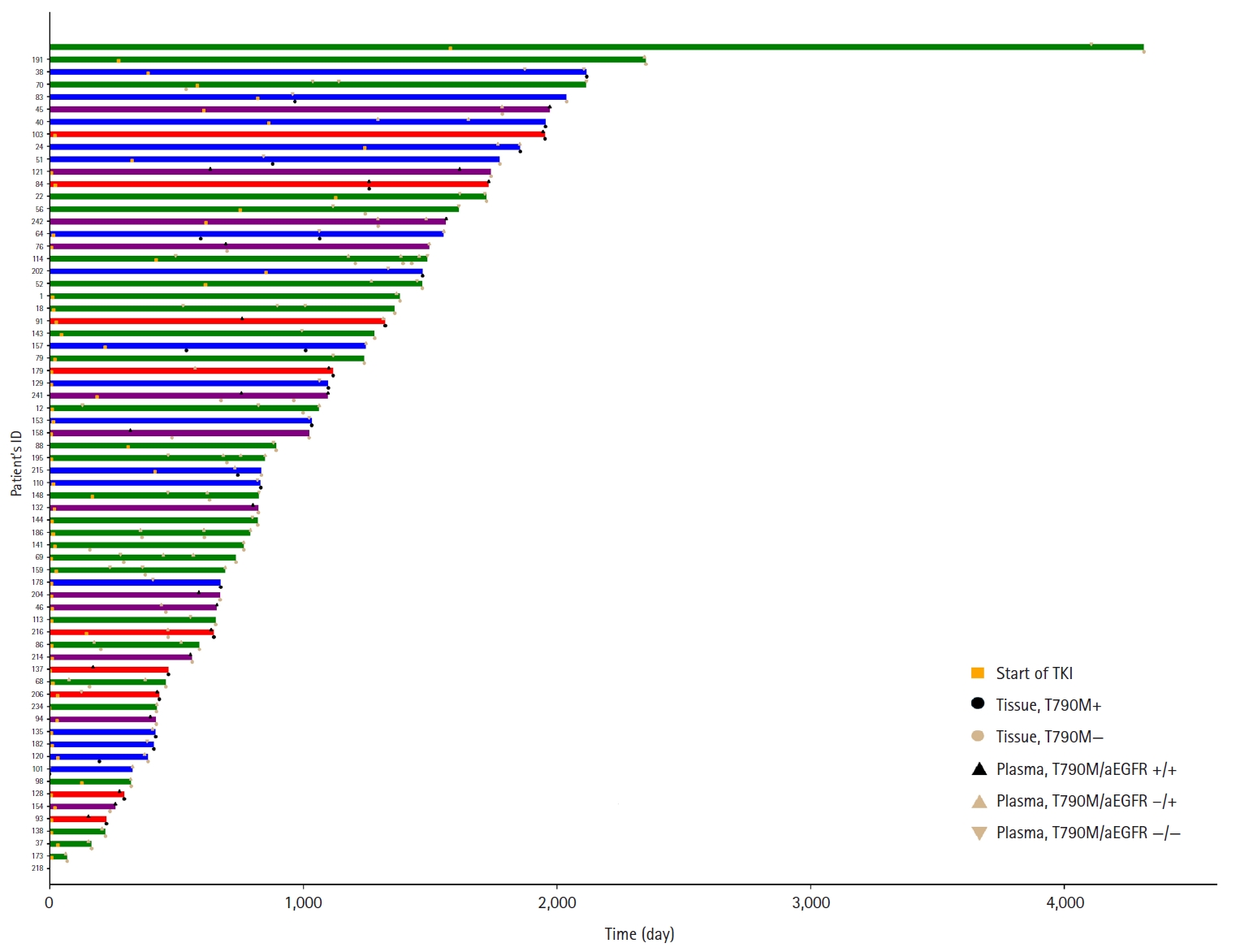

Fig. 1.
Graphical abstract
| Parameter | Value |
|---|---|
| Age (yr) | 67 (40–86) |
| Sex | |
| Male | 106 (42.7) |
| Female | 142 (57.3) |
| Pathologic diagnosis | |
| Adenocarcinoma | 238 (96.0) |
| Squamous cell carcinoma | 8 (3.2) |
| Adenosquamous carcinoma | 2 (0.8) |
| Stage | |
| I | 16 (6.5) |
| II | 10 (4.0) |
| III | 39 (15.7) |
| IV | 183 (73.8) |
| Tissue | Plasma | |
|---|---|---|
| Activating mutation | ||
| Exon 19 deletion | 139 (56.0) | 102 (41.1) |
| L858R | 87 (35.1) | 53 (21.4) |
| L861Q | 4 (1.6) | 5 (2.0) |
| S768I | 1 (0.4) | 0 |
| G719C | 1 (0.4) | 0 |
| T790M | 1 (0.4) | 0 |
| Exon 20 insertion | 4 (1.6) | 1 (0.4) |
| G719X/L861Q | 3 (1.2) | 1 (0.4) |
| 19del/L861Q | 2 (0.8) | 0 |
| L858R/ T790M | 1 (0.4) | 0 |
| S768I/G719S | 1 (0.4) | 0 |
| L858R/S768I | 1 (0.4) | 1 (0.4) |
| Negative | 3 (1.2) | 85 (34.3) |
| Total | 248 | 248 |
| Resistance mutation | ||
| T790M | 28 (40.6) | 57 (27.5) |
| Negative | 41 (59.4) | 150 (72.5) |
| Total | 69 | 207 |
| Activating EGFR mutation | T790M mutation | |||||
|---|---|---|---|---|---|---|
| Tissue | p-value | Tissue | p-value | |||
| Positive | Negative | Positive | Negative | |||
| Plasma | ||||||
| Positive | 159 (65.4) | 4 (1.6) | .371 | 9 (13.4) | 12 (17.9) | .898 |
| Negative | 85 (31.6) | 0 | 17 (25.4) | 28 (43.3) | ||
| Activating mutation | p-value | T790M | p-value | |||
|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |||
| T category | ||||||
| T1, T2 | 71 | 50 | .052 | 27 | 75 | .901 |
| T3, T4 | 88 | 35 | 29 | 74 | ||
| N category | ||||||
| N0 | 34 | 34 | .013 | 42 | 111 | .992 |
| N1–N3 | 124 | 50 | 14 | 37 | ||
| M category | ||||||
| M0 | 30 | 35 | .012 | 13 | 45 | .299 |
| M1 | 133 | 50 | 44 | 5 | ||
Values are presented as mean (range) or number (%). SCC, squamous cell carcinoma.
Values are presented as number (%). EGFR, epidermal growth factor receptor.
Values are presented as number (%). EGFR, epidermal growth factor receptor.
EGFR, epidermal growth factor receptor.

 E-submission
E-submission