Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 50(6); 2016 > Article
-
Original Article
CD9 Expression in Colorectal Carcinomas and Its Prognostic Significance - Kyung-Ju Kim, Hee Jung Kwon, Min Chong Kim, Young Kyung Bae
-
Journal of Pathology and Translational Medicine 2016;50(6):459-468.
DOI: https://doi.org/10.4132/jptm.2016.10.02
Published online: October 25, 2016
Department of Pathology, Yeungnam University College of Medicine, Daegu, Korea
- Corresponding Author Young Kyung Bae, MD Department of Pathology, Yeungnam University College of Medicine, 170 Hyeonchung-ro, Nam-gu, Daegu 42415, Korea Tel: +82-53-640-6755 Fax: +82-53-622-8432 E-mail: ykbae@ynu.ac.kr
© 2016 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- CD9, a member of the tetraspanin superfamily, is a tumor suppressor in many malignancies. The aim of this study was to evaluate the immunohistochemical expression of CD9 in colorectal carcinomas (CRCs) and determine clinicopathological and prognostic significance of its expression.
-
Methods
- The CD9 expression status of 305 CRCs was evaluated using a semi-quantitative scoring system in tumor cells (T-CD9) and immune cells (I-CD9) by classifying the results as high and low expression.
-
Results
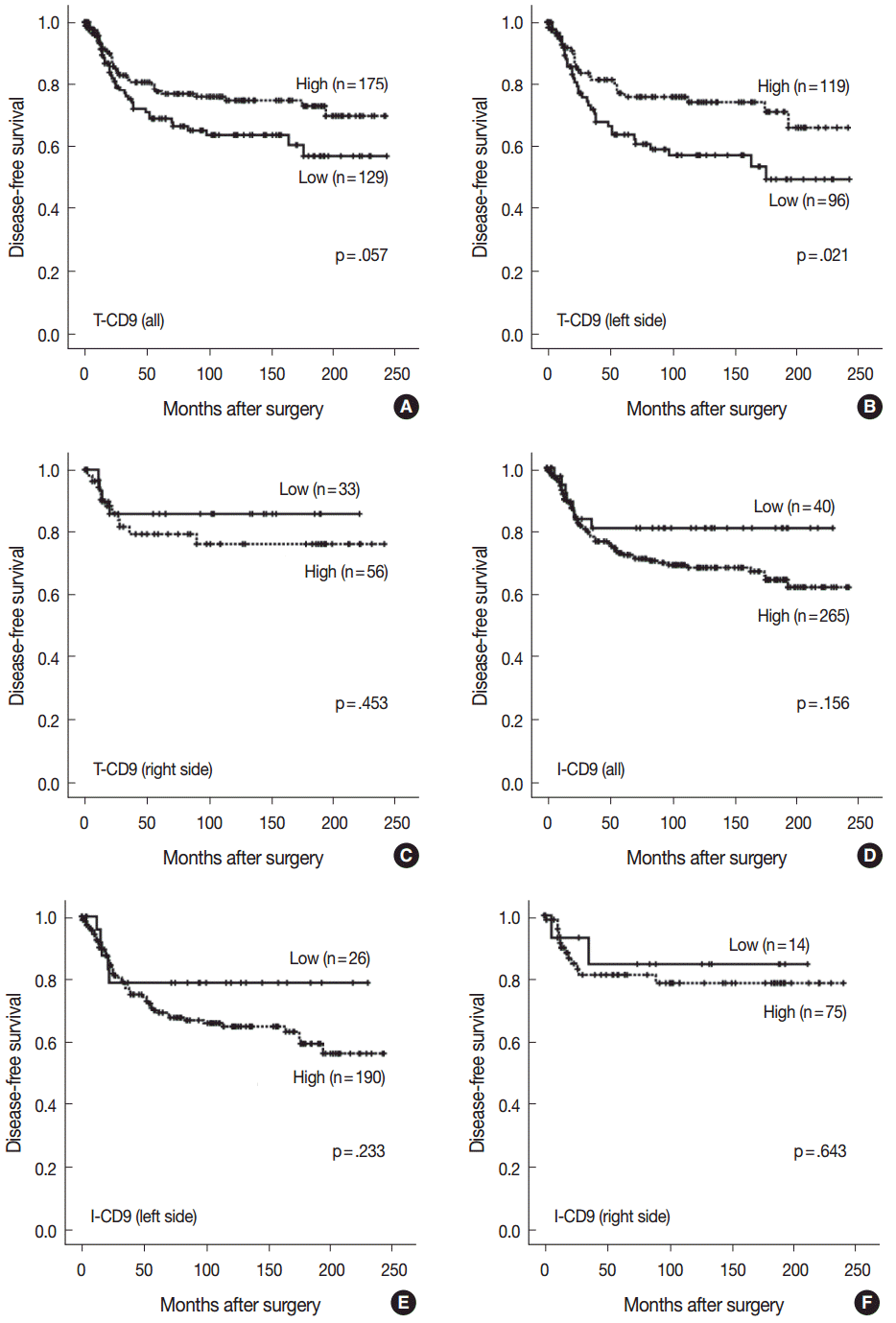
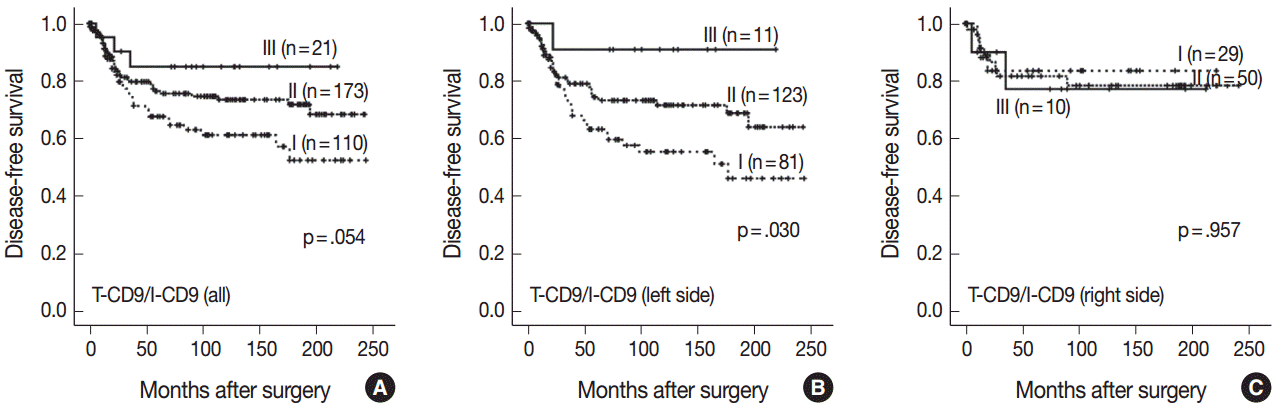
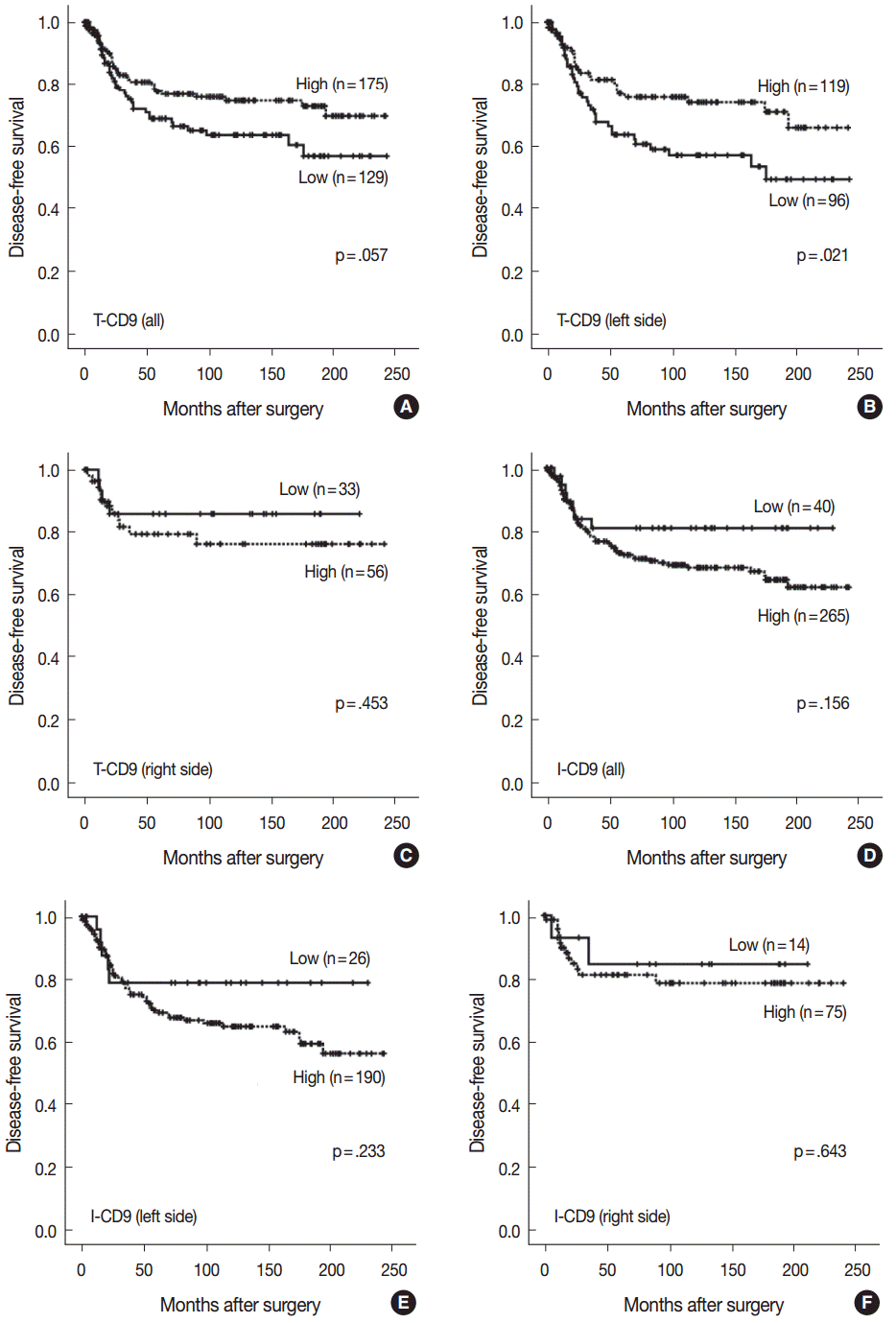
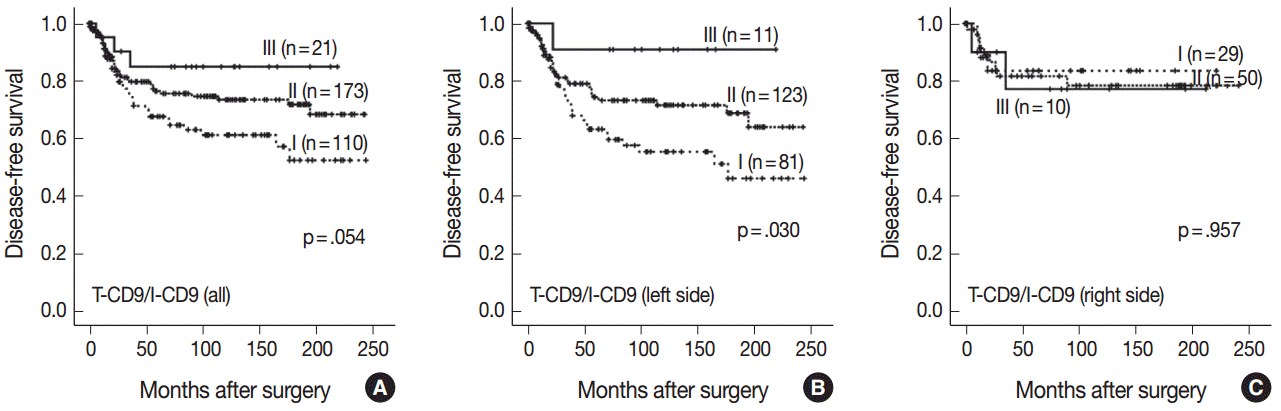
- High T-CD9 (T-CD9 [+]) expression was detected in 175 samples (57.6%) and high I-CD9 (I-CD9 [+]) expression was detected in 265 samples (86.9%). Using Kaplan-Meier survival analysis, the T-CD9 (+) group showed a tendency for better disease-free survival (DFS) (p = .057). In left-sided tumors, DFS was significantly longer in the T-CD9 (+) group (p = .021) but no statistical significance was observed with right-sided tumors (p = .453). I-CD9 (+) CRCs significantly correlated with well/moderately differentiation (p = .014). In Kaplan-Meier analysis, the I-CD9 (+) group had a tendency towards worse DFS compared to the I-CD9 (–) group (p = .156). In combined survival analysis of T-CD9 and I-CD9, we found that the longest DFS was among patients in the T-CD9 (+)/I-CD9 (–) group, whereas the T-CD9 (–)/I-CD9 (+) group showed the shortest DFS (p = .054).
-
Conclusions
- High expression of T-CD9 was associated with a favorable DFS, especially in left-sided CRCs. Combined evaluation of T-CD9 and I-CD9 is required to determine the comprehensive prognostic effect of CD9 in CRCs.
- Patients and specimens
- A total of 354 CRC formalin-fixed paraffin-embedded tissues were collected retrospectively from the pathologic archives of Yeungnam University Medical Center, Daegu, Korea. All samples were derived from patients who underwent radical resection and subsequent adjuvant chemotherapy from 1996 to 2000 in our institution. Among these, 49 patients (32 patients who had preoperative neoadjuvant chemoradiation therapy, 16 patients who were diagnosed with stage IV disease, and 1 patient who was diagnosed with stage IV and had preoperative neoadjuvant chemoradiation therapy at the same time) were excluded. Patients’ electronic medical records and pathologic reports were reviewed to obtain information including age, sex, location of tumor, pT/pN categories, presence of lymphatic/venous/perineural invasion, and tumor differentiation. The follow-up period for patients (from the date of surgery to the date of recurrence or the last follow-up) ranged from 4 to 243 months and the average disease-free survival (DFS) was 85 months. The restaging of tumors and histological re-grading were carried out according to the American Joint Committee on Cancer Staging Manual, seventh edition. The study was approved by the Yeungnam University Medical Center Institutional Review Board (No. 2016-07-020).
- Tissue microarray and immunohistochemistry
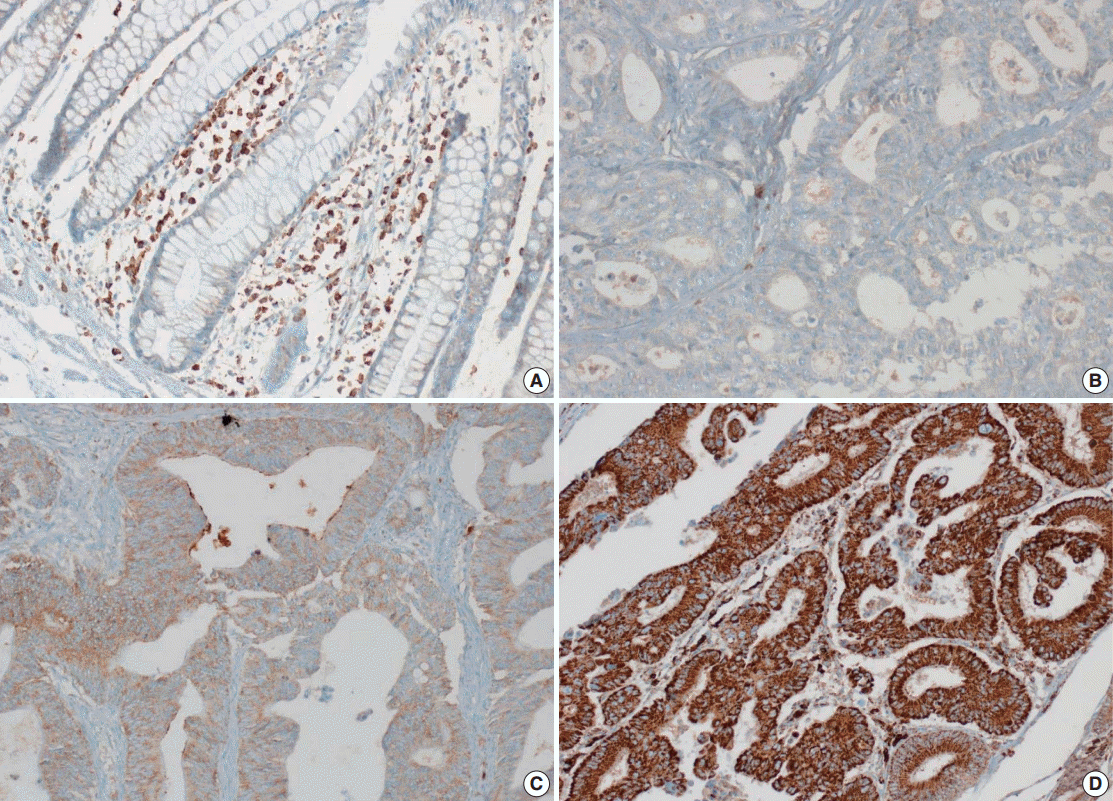
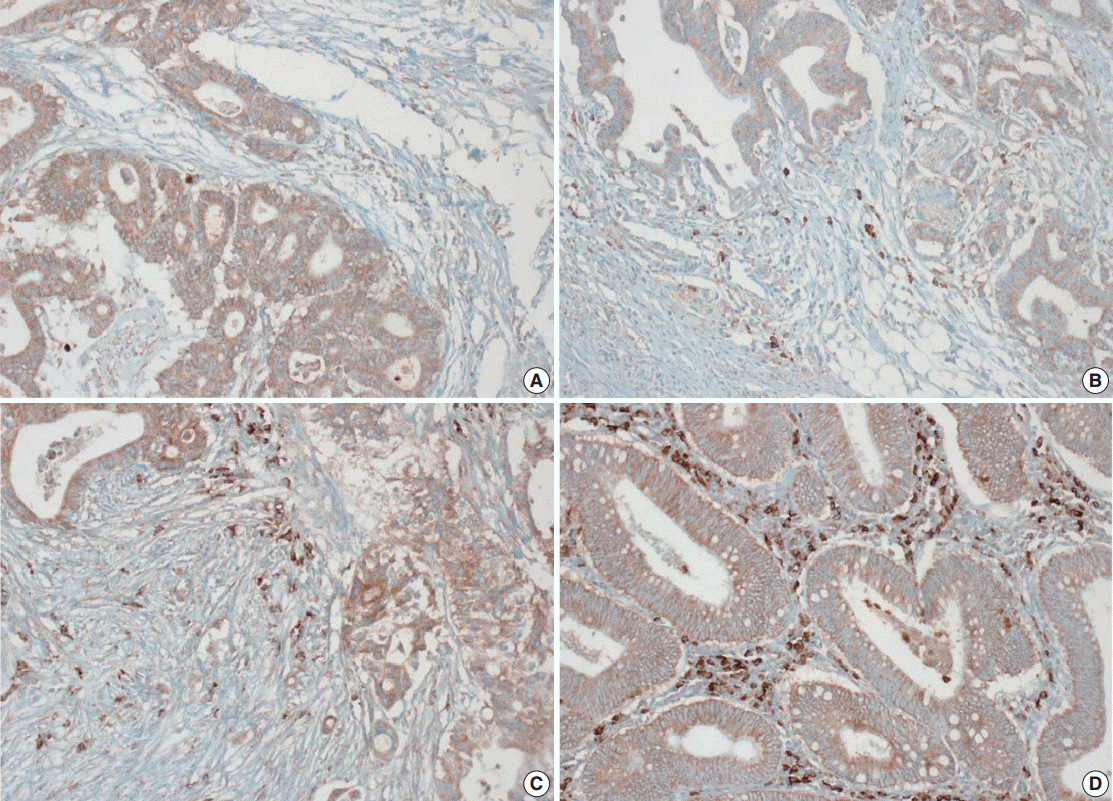
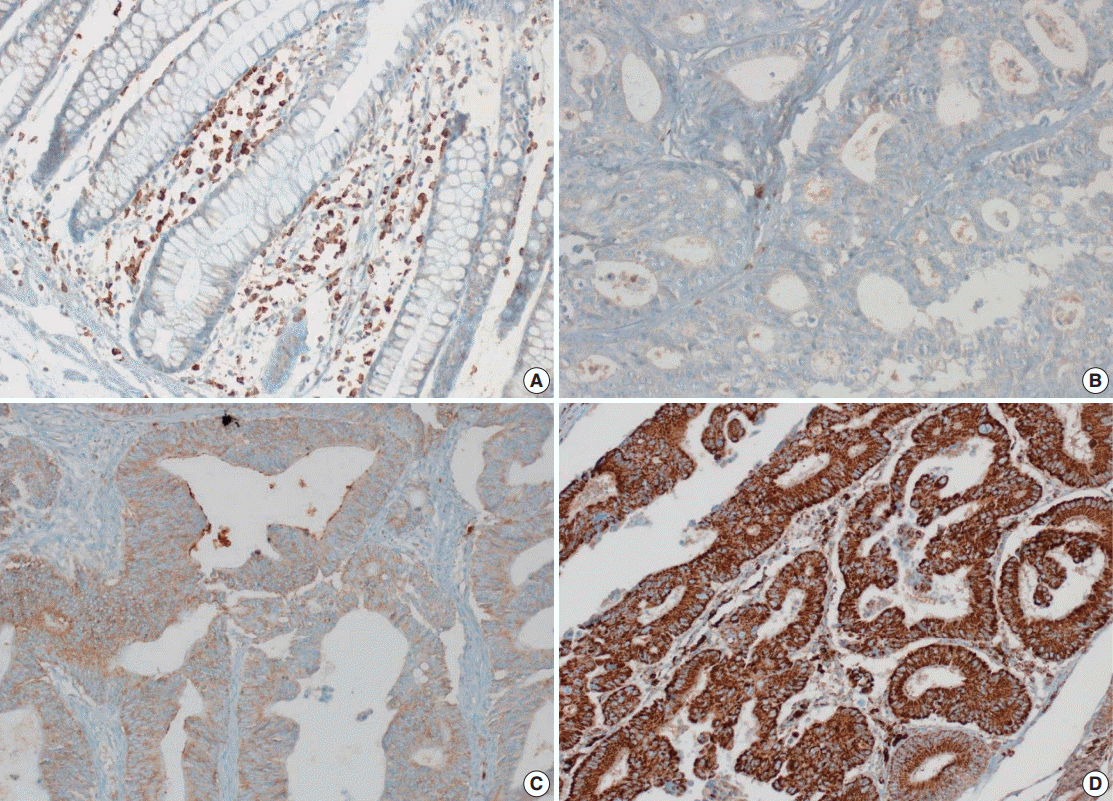
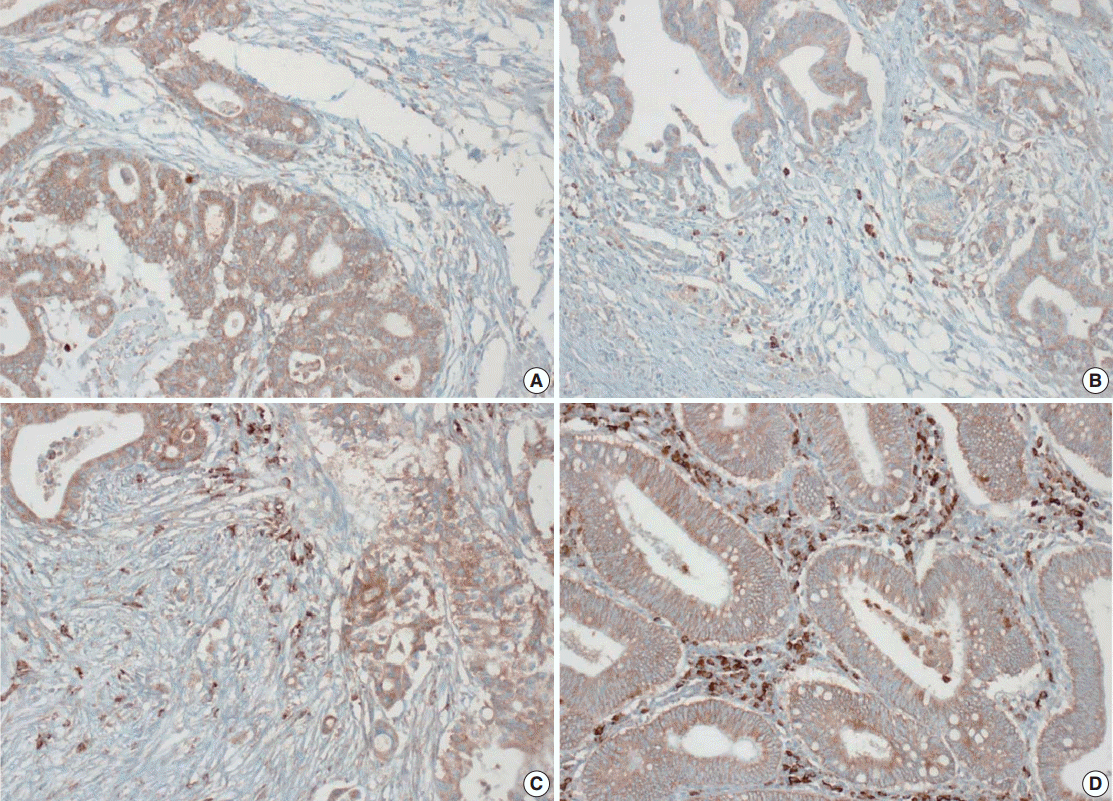
- Tissue microarray (TMA) construction was conducted by reviewing hematoxylin and eosin stained slides and selecting an area that had sufficient tumor with no hemorrhage or necrosis. One representative tissue core (2 mm in diameter) was obtained from a donor block and placed in a recipient block using a trephine apparatus. Five cases of non-neoplastic colorectal (Fig. 1A), renal, hepatic parenchymal tissues and three cases of non-neoplastic gastric parenchymal tissues were included in each TMA. Seven tissue array blocks were prepared for 305 cases. Immunohistochemical (IHC) staining with rabbit monoclonal Abs against CD9 (1:1,200, EPR2949; Abcam, Cambridge, UK) was performed on each TMA block using a BenchMark XT immunostainer (Ventana Medical System, Tucson, AZ, USA) following the manufacturer’s protocol. Immunoreactivity of CD9 expression was assessed in tumor cells (T-CD9) and immune cells (I-CD9) in stroma semiquantitatively. In cases of T-CD9, both intensity and extent of membranous-to-cytoplasmic staining were evaluated. The intensity was initially scored on a scale of 0 to 3: negative (0), weak positive (1+), moderately positive (2+), and strongly positive (3+) (Fig. 1B–D). The extent of staining was categorized into five groups according to the percentages of positively stained cells: 0% (0), ≤25% (1+), >25 and ≤50% (2+), >50 and ≤75% (3+), and >75% (4+). The final T-CD9 score was determined by multiplying the extent and intensity scores and recording the final score from 0 to 12. The T-CD9 immunoreactivity score was then dichotomized as low (score, 0 to 3) or high (score, 4 to 12). In cases of I-CD9, the percentage of stained immune cells was estimated on a 0, 1+, 2+, and 3+ scale: 0% (0), ≤25% (1+), >25 and ≤ 50% (2+), and >50% (3+) (Fig. 2). The cases of I-CD9 score 0 were classified as low, and when the score was 1 to 3, they were classified as high. All IHC staining assessments were performed independently by two pathologists (K.-J.K. and Y.K.B.), who were blinded to the clinicopathological information. Conflicting evaluations between the pathologists were discussed and a consensus was achieved.
- Statistical analysis
- IBM SPSS ver. 20.0 (IBM Corp., Armonk, NY, USA) was used for all statistical analysis. Correlation between the categorical variables was analyzed using Pearson’s chi-square test or Fisher exact test. DFS was defined as the period from the date of primary radical resection to the date of tumor recurrence, metastasis, death, or the last follow-up. DFS was assessed by the Kaplan-Meier method with the log-rank test, and multivariate analysis with the Cox regression model was used to adjust variables that had been statistically significant for DFS in univariate analysis. All statistical analyses were two-sided, and statistical significance was considered to be p<.05.
MATERIALS AND METHODS
- CD9 expression status in CRC
- Five normal sample of colorectal tissue and 305 CRC samples were examined to identify CD9 expression. Of the 305 CRC samples, one case T-CD9 sample was excluded from evaluation because repeated sectioning of the TMA block resulted in very few tumor cells remaining for evaluation. As a result, 304 cases of T-CD9 and 305 cases of I-CD9 were analyzed. Among the five samples of non-neoplastic colorectal mucosae, the expression of CD-9 was negative or weakly positive in almost all epithelial cells (Fig. 1A). These weakly positive cells showed similar intensity and were classified into the low expression group. On the other hand, more than 50% of stained stromal immune cells in non-neoplastic colorectal tissues exhibited strong expression of CD9 throughout the entire samples and were classified as the high expression group. Meanwhile, tumor cells and surrounding stromal immune cells showed variable expression patterns. T-CD9 (+) was detected in 175 of 304 CRCs (57.6%), and I-CD9 (+) was detected in 265 of 305 cases (86.9%). One hundred fifty-four cases (50.7%) showed both T-CD9 (+) and I-CD9 (+) and 19 cases (6.3%) exhibited both T-CD9 (–) and I-CD9 (–).
- Characteristics of a T-CD9 subset of CRC
- No statistically significant differences were noted in clinicopathological parameters including age, sex, stage, gross type, location, pT/pN categories, or frequency of angiolymphatic and perineural invasion between the T-CD9 (+) and T-CD9 (–) groups (Table 1). In Kaplan-Meier survival analysis, the T-CD9 (+) group showed a tendency for better DFS than the T-CD9 (–) group, but statistical significance was not reached (p=.057) (Fig. 3A). We additionally subdivided cases into two groups according to tumor location: right-sided tumors (n=89) and left-sided tumors (n=215). Based on the previous literature [18], CRCs were classified into right- and left-sided tumors relative to the splenic flexure. In other words, tumors arising proximal to the splenic flexure (cecum, ascending colon, and transverse colon) are considered right-sided, and tumors distal to the splenic flexure (descending colon, sigmoid colon, and rectum) are considered left-sided. In left-sided tumors, DFS was significantly better in the T-CD9 (+) group compared to the T-CD9 (–) group (p=.021) (Fig. 3B). However, in right-sided tumors, no significant difference was observed between the two groups (p=.453) (Fig. 3C). Multivariate analysis including stage, lymphatic invasion, vascular invasion, perineural invasion and T-CD9—which were significant prognostic factors in univariate analysis—revealed that the stage (p=.002) was still an independent prognostic predictor for DFS, whereas T-CD9 expression failed to correlate with prognosis, with no difference between T-CD9 (+) and T-CD9 (–) groups in patients with left-sided CRC (p=.167) (Table 2).
- Characteristics of an I-CD9 subset of CRC
- I-CD9 (+) CRCs significantly correlated with well/moderately differentiation (p=.014) compared to I-CD9 (–) CRCs. However, no significant associations were found for other parameters including age, sex, stage, gross type, location, pT/pN categories, or frequency of angiolymphatic and perineural invasion (Table 1). In Kaplan-Meier survival analysis, the I-CD9 (+) group had a tendency towards worse DFS compared to the I-CD9 (–) group, but no significant difference was found (p=.156) (Fig. 3D). When the cases were subdivided into left- and right-sided CRC groups, I-CD9 (–) suggested a potential protective role in left-sided tumors (p=.233), but statistical significance was not reached (Fig. 3E). In right-sided tumors, no statistical significance was observed between the two groups (p=.643) (Fig. 3F).
- Characteristics of a combined T-CD9/I-CD9 subset of CRC
- Kaplan-Meier survival analysis found the longest DFS for patients in the T-CD9 (+)/I-CD9 (–) group, whereas the T-CD9 (–)/I-CD9 (+) group had the shortest DFS, but statistical significance was not reached (p=.054) (Fig. 4A). When the patients were divided into left- and right-sided CRC groups, increased survival differences between subgroups was observed in left-sided tumors (p=.030) (Fig. 4B). In right-sided tumors, no significant difference in DFS was observed (p=.957) (Fig. 4C). In multivariate analysis using a Cox proportional hazards model, combined T-CD9/I-CD9 expression was not an independent prognostic factor of DFS in left-sided CRCs (p=.199) (Table 2).
RESULTS
- Many previous in vitro and in vivo models of cancer cell lines have contributed to our understanding of the role of CD9 and the associated functional complexes with other members of the tetraspanin family and various integrins [14-16,19]. High expression of CD9 in tumor cells has been associated with favorable prognosis, reduced metastatic potential, and cancer growth in various tumor types, including lung, breast, prostate, stomach, pancreas, and colon carcinoma [4,20-22]. Miyake et al. [23] demonstrated that injection of CD9-transfected melanoma cells suppressed lung metastasis in a rodent model and Ovalle et al. [16] showed that ectopic CD9 expression in a colon cancer cell line resulted in inhibition of cell growth. In addition, Mori et al. [24] stated that reduced expression of CD9 by reverse transcription polymerase chain reaction is linked to poor prognosis in colon cancer. Our data demonstrated that patients with high expression of CD9 in tumor cells (T-CD9 [+]) tended to have a longer DFS, although the difference was not statistically significant (p=.057).
- However, studies have yielded controversial results about CD9 expression in non-neoplastic colorectal cells. Mori et al. [24] demonstrated that CD9 mRNA is consistently expressed in non-neoplastic colonic tissues adjacent to tumors and its expression level was higher in normal colonic tissues than in tumors in six normal/tumor paired samples using northern blot analysis. Okochi et al. [25] also showed that strong CD9 expression was detected in normal colonic epithelium by IHC. On the contrary, negative or weakly positive expression of CD9 was observed in normal colorectal epithelial cells in our study. These contradictory finding of the non-neoplastic CD9 expression may be attributed to differing experimental techniques such as northern blot analysis and IHC, as well as variation in the different types of CD9 antibodies. Furthermore, our data has limitation in this regard, because very few normal colorectal tissue samples (n=5) were included, and pairing of normal and tumor samples has not been carried out. Further investigation using extended cases of paired samples are needed for clarification.
- Several mechanisms have been suggested to explain the tumor suppressant function of CD9 [4,26,27]. In conjunction with EGFR, CD9 has been shown to induce apoptosis via restricted activation of p46 Shc isoforms or the c-Jun N-terminal kinase/stress-activated protein kinase and p38 mitogen-activated protein kinase pathways. By forming a functional complex with integrin β1, CD9 suppresses cell motility and enhances cell integrity and adhesion. Furthermore, CD9 may hinder cell proliferation through internalization of EGFR, resulting in attenuation of EGF-EGFR induced signals. Notably, the protective prognostic role of T-CD9 on DFS was demonstrated in the relatively homogenous left-sided CRC group in this study with statistical significance (p=.021). However, in the right-sided tumor group, no significance of T-CD9 was observed in DFS, but this finding was considered to have limited reliability due to the small number of cases (n=89). The prognostic effect of T-CD9 is speculated to be attributed to different molecular carcinogenic mechanisms between right- and left-sided CRCs [28]. In left-sided CRCs, the majority of tumors develop through a chromosomal instability pathway known as the adenoma-carcinoma sequence. In this pathway, progression to adenoma-carcinoma from normal mucosa is established by accumulation of various genomic changes including activation of proto-oncogenes such as KRAS and inactivation of tumor suppressor genes such as APC and TP53 [29]. Meanwhile, a large proportion of tumors with microsatellite unstable (microsatellite instability–unstable) subtypes arise predominantly in the right side of the colon. They are caused by genetic and epigenetic alterations of mismatch repair genes and consequently exhibit a hypermutated phenotype. Serrated morphology tumors and CpG island methylator phenotypes are seen more frequently in right-sided tumors than in left-sided CRCs. These findings suggest that even in the same tumor, the prognostic implication of CD9 can be different depending on the underlying molecular mechanisms of carcinogenesis. Extended study is required to verify the hypothesis presented above and the location based variation of CD9 roles in tumor progression.
- Tumor-infiltrating immune cells are thought to play a substantial role in shaping the microenvironment depending on various factors such as cytokines and chemokines, towards either immunostimulatory antitumor conditions or immunoregulatory tumor promoting mileu [17]. In survival analysis, I-CD9 (+) showed a tendency to be associated with a high recurrence rate in left-sided CRCs (p=.156) and this finding is contrary to the case of T-CD9. In a previous study, Erovic et al. [6] described the CD9-positive peritumoral lymphocytes in squamous cell carcinoma of the head and neck. Bruno et al. [30] demonstrated that CD9-expressing tumor-infiltrating immune cells, especially tumor-associated natural killer cells, are polarized to have pro-angiogenic and pro-tumoral phenotypes. These findings are consistent with our results regarding I-CD9. To the best of our knowledge, this is the first report to identify a differential prognostic role of CD9 expression in tumor cells and immune cells in CRCs.
- Murayama et al. [4] stated that a therapeutic agent targeting tetraspanin, especially CD9, using monoclonal Abs such as ALB6 and PAIN13, might be beneficial for cancer patients. However, several researchers have suggested the contradictory finding of T-CD9 in various cancer types including multiple myeloma [31], fibrosarcoma [32], and even in gastric carcinoma [25,26], showing that CD9 expression of tumor cells has a positive influence on tumor proliferation and invasion by enhancing HB-EGF/EGFR interaction or metalloproteinases-2 secretion. In addition, our study also demonstrated an opposite effect of T-CD9 and I-CD9 on tumor progression. Therefore, future studies should focus on the varying mechanisms and the therapeutic response of I-CD9 and T-CD9 in CRCs.
- In combined survival analysis of T-CD9 and I-CD9, tumors with high T-CD9 expression plus low I-CD9 expression showed a tendency to have a longer DFS compared with (1) tumors with concurrent high or low expression of T-CD9 and I-CD9, or (2) tumors with low T-CD9 expression plus high I-CD9 expression (p=.054), and a similar result was found in left-sided CRCs with statistical significance (p=.030). These findings suggested that the combined evaluation of T-CD9 and I-CD9 is required to determine the comprehensive prognostic effect of CD9 in CRCs.
- In conclusion, high expression of CD9 in tumor cells tends to be inversely related to tumor recurrence, especially in left-sided CRCs. However, a different tendency on DFS was demonstrated in immune cells regardless of tumor location. Additional studies are necessary, with an emphasis on differential expression of CD9 in tumor cells and immune cells.
DISCUSSION
Acknowledgments
| Variable |
CD9 expression in tumor cells (T-CD9)a |
CD9 expression in immune cells (I-CD9) |
||||||
|---|---|---|---|---|---|---|---|---|
| No. of cases | Low | High | p-value | No. of cases | Low | High | p-value | |
| Age (yr) | .449 | .496 | ||||||
| ≤ 60 | 204 | 83 (64.3) | 121 (69.1) | 205 | 25 (62.5) | 180 (67.9) | ||
| > 60 | 100 | 46 (35.7) | 54 (30.9) | 100 | 15 (37.5) | 85 (32.1) | ||
| Sex | .175 | .581 | ||||||
| Male | 163 | 75 (58.1) | 88 (50.3) | 163 | 23 (57.5) | 140 (52.8) | ||
| Female | 141 | 54 (41.9) | 87 (49.7) | 142 | 17 (42.5) | 125 (47.2) | ||
| Tumor location | .224 | .385 | ||||||
| Right | 89 | 33 (25.6) | 56 (32.0) | 89 | 14 (35.0) | 75 (28.3) | ||
| Left | 215 | 96 (74.4) | 119 (68.0) | 216 | 26 (65.0) | 190 (71.7) | ||
| Gross typeb | .775 | .632 | ||||||
| Fungating | 137 | 61 (48.0) | 76 (46.3) | 137 | 16 (43.2) | 121 (47.5) | ||
| Infiltrative | 154 | 66 (52.0) | 88 (53.7) | 155 | 21 (56.8) | 134 (52.5) | ||
| AJCC TNM stage | .938 | .156 | ||||||
| Stage I | 32 | 13 (10.1) | 19 (10.8) | 33 | 1 (2.5) | 32 (12.1) | ||
| Stage II | 134 | 56 (43.4) | 78 (44.6) | 134 | 21 (52.5) | 113 (42.6) | ||
| Stage III | 138 | 60 (46.5) | 78 (44.6) | 138 | 18 (45.0) | 120 (45.3) | ||
| Tumor depth (pT) | .909 | .137 | ||||||
| pT1 | 9 | 4 (3.1) | 5 (2.8) | 9 | 0 | 9 (3.4) | ||
| pT2 | 30 | 11 (8.5) | 19 (10.9) | 31 | 1 (2.5) | 30 (11.3) | ||
| pT3 | 233 | 101 (78.3) | 132 (75.4) | 233 | 36 (90.0) | 197 (74.3) | ||
| pT4 | 32 | 13 (10.1) | 19 (10.9) | 32 | 3 (7.5) | 29 (11.0) | ||
| LN metastasis (pN) | .884 | .935 | ||||||
| pN0 | 167 | 69 (53.5) | 98 (56.0) | 168 | 22 (55.0) | 146 (55.1) | ||
| pN1 | 86 | 37 (28.7) | 49 (28.0) | 86 | 12 (30.0) | 74 (27.9) | ||
| pN2 | 51 | 23 (17.8) | 28 (16.0) | 51 | 6 (15.0) | 45 (17.0) | ||
| Tumor differentiationb | .734 | .014 | ||||||
| WD/MD | 261 | 110 (85.9) | 151 (87.3) | 262 | 29 (74.4) | 233 (88.6) | ||
| PD/Other | 40 | 18 (14.1) | 22 (12.7) | 40 | 10 (25.6) | 30 (11.4) | ||
| Lymphatic invasionb | .394 | .361 | ||||||
| Absent | 192 | 77 (67.5) | 115 (72.3) | 193 | 21 (63.6) | 172 (71.4) | ||
| Present | 81 | 37 (32.5) | 44 (27.7) | 81 | 12 (36.4) | 69 (28.6) | ||
| Venous invasionb | .467 | .300 | ||||||
| Absent | 208 | 85 (74.6) | 123 (78.3) | 209 | 23 (69.7) | 186 (77.8) | ||
| Present | 63 | 29 (25.4) | 34 (21.7) | 63 | 10 (30.3) | 53 (22.2) | ||
| Perineural invasionb | .553 | .856 | ||||||
| Absent | 195 | 84 (75.0) | 111 (78.2) | 196 | 25 (78.1) | 171 (76.7) | ||
| Present | 59 | 28 (25.0) | 31 (21.8) | 59 | 7 (21.9) | 52 (23.3) | ||
Values are presented as number (%).
AJCC, American Joint Committee on Cancer; LN, lymph node; WD, well differentiated; MD, moderately differentiated; PD, poorly differentiated.
aOne case is excluded from this statistical analysis due to unavailable TMA data;
bOnly patients with available clinicopathological data are included.
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87-108. ArticlePubMedPDF
- 2. Dalerba P, Maccalli C, Casati C, Castelli C, Parmiani G. Immunology and immunotherapy of colorectal cancer. Crit Rev Oncol Hematol 2003; 46: 33-57. ArticlePubMed
- 3. Kim Y, Bae JM, Li G, Cho NY, Kang GH. Image analyzer-based assessment of tumor-infiltrating T cell subsets and their prognostic values in colorectal carcinomas. PLoS One 2015; 10: e0122183. Article
- 4. Murayama Y, Oritani K, Tsutsui S. Novel CD9-targeted therapies in gastric cancer. World J Gastroenterol 2015; 21: 3206-13. ArticlePubMedPMC
- 5. Le Naour F, André M, Greco C, et al. Profiling of the tetraspanin web of human colon cancer cells. Mol Cell Proteomics 2006; 5: 845-57. ArticlePubMed
- 6. Erovic BM, Pammer J, Hollemann D, et al. Motility-related protein-1/CD9 expression in head and neck squamous cell carcinoma. Head Neck 2003; 25: 848-57. ArticlePubMed
- 7. Regina Todeschini A, Hakomori SI. Functional role of glycosphingolipids and gangliosides in control of cell adhesion, motility, and growth, through glycosynaptic microdomains. Biochim Biophys Acta 2008; 1780: 421-33. ArticlePubMed
- 8. Kwon HJ, Min SY, Park MJ, et al. Expression of CD9 and CD82 in clear cell renal cell carcinoma and its clinical significance. Pathol Res Pract 2014; 210: 285-90. ArticlePubMed
- 9. Huang CI, Kohno N, Ogawa E, Adachi M, Taki T, Miyake M. Correlation of reduction in MRP-1/CD9 and KAI1/CD82 expression with recurrences in breast cancer patients. Am J Pathol 1998; 153: 973-83. ArticlePubMedPMC
- 10. Chen Z, Gu S, Trojanowicz B, et al. Down-regulation of TM4SF is associated with the metastatic potential of gastric carcinoma TM4SF members in gastric carcinoma. World J Surg Oncol 2011; 9: 43.ArticlePubMedPMCPDF
- 11. Houle CD, Ding XY, Foley JF, Afshari CA, Barrett JC, Davis BJ. Loss of expression and altered localization of KAI1 and CD9 protein are associated with epithelial ovarian cancer progression. Gynecol Oncol 2002; 86: 69-78. ArticlePubMed
- 12. Furuya M, Kato H, Nishimura N, et al. Down-regulation of CD9 in human ovarian carcinoma cell might contribute to peritoneal dissemination: morphologic alteration and reduced expression of beta1 integrin subsets. Cancer Res 2005; 65: 2617-25. PubMed
- 13. Yin M, Soikkeli J, Jahkola T, Virolainen S, Saksela O, Hölttä E. Osteopontin promotes the invasive growth of melanoma cells by activating integrin alphavbeta3 and down-regulating tetraspanin CD9. Am J Pathol 2014; 184: 842-58. PubMed
- 14. Ikeyama S, Koyama M, Yamaoko M, Sasada R, Miyake M. Suppression of cell motility and metastasis by transfection with human motility-related protein (MRP-1/CD9) DNA. J Exp Med 1993; 177: 1231-7. ArticlePubMedPMCPDF
- 15. Takeda T, Hattori N, Tokuhara T, Nishimura Y, Yokoyama M, Miyake M. Adenoviral transduction of MRP-1/CD9 and KAI1/CD82 inhibits lymph node metastasis in orthotopic lung cancer model. Cancer Res 2007; 67: 1744-9. ArticlePubMedPDF
- 16. Ovalle S, Gutiérrez-López MD, Olmo N, et al. The tetraspanin CD9 inhibits the proliferation and tumorigenicity of human colon carcinoma cells. Int J Cancer 2007; 121: 2140-52. ArticlePubMedPDF
- 17. Salgado R, Denkert C, Demaria S, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol 2015; 26: 259-1. PubMed
- 18. Iacopetta B. Are there two sides to colorectal cancer? Int J Cancer 2002; 101: 403-8. ArticlePubMed
- 19. Lee JH, Bae JA, Lee JH, et al. Glycoprotein 90K, downregulated in advanced colorectal cancer tissues, interacts with CD9/CD82 and suppresses the Wnt/beta-catenin signal via ISGylation of betacatenin. Gut 2010; 59: 907-17. ArticlePubMed
- 20. Sho M, Adachi M, Taki T, et al. Transmembrane 4 superfamily as a prognostic factor in pancreatic cancer. Int J Cancer 1998; 79: 509-16. ArticlePubMed
- 21. Boucheix C, Duc GH, Jasmin C, Rubinstein E. Tetraspanins and malignancy. Expert Rev Mol Med 2001; 2001: 1-17. Article
- 22. Hashida H, Takabayashi A, Tokuhara T, et al. Integrin alpha3 expression as a prognostic factor in colon cancer: association with MRP-1/CD9 and KAI1/CD82. Int J Cancer 2002; 97: 518-25. PubMed
- 23. Miyake M, Inufusa H, Adachi M, et al. Suppression of pulmonary metastasis using adenovirally motility related protein-1 (MRP-1/CD9) gene delivery. Oncogene 2000; 19: 5221-6. ArticlePubMedPDF
- 24. Mori M, Mimori K, Shiraishi T, et al. Motility related protein 1 (MRP1/CD9) expression in colon cancer. Clin Cancer Res 1998; 4: 1507-10. PubMed
- 25. Okochi H, Mine T, Nashiro K, Suzuki J, Fujita T, Furue M. Expression of tetraspans transmembrane family in the epithelium of the gastrointestinal tract. J Clin Gastroenterol 1999; 29: 63-7. ArticlePubMed
- 26. Hori H, Yano S, Koufuji K, Takeda J, Shirouzu K. CD9 expression in gastric cancer and its significance. J Surg Res 2004; 117: 208-15. ArticlePubMed
- 27. Soyuer S, Soyuer I, Unal D, Ucar K, Yildiz OG, Orhan O. Prognostic significance of CD9 expression in locally advanced gastric cancer treated with surgery and adjuvant chemoradiotherapy. Pathol Res Pract 2010; 206: 607-10. ArticlePubMed
- 28. Bae JM, Kim JH, Kang GH. Molecular subtypes of colorectal cancer and their clinicopathologic features, with an emphasis on the serrated neoplasia pathway. Arch Pathol Lab Med 2016; 140: 406-12. ArticlePubMedPDF
- 29. Armaghany T, Wilson JD, Chu Q, Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res 2012; 5: 19-27. PubMedPMC
- 30. Bruno A, Ferlazzo G, Albini A, Noonan DM. A think tank of TINK/TANKs: tumor-infiltrating/tumor-associated natural killer cells in tumor progression and angiogenesis. J Natl Cancer Inst 2014; 106: dju200.ArticlePubMedPMC
- 31. Wang YD, De Vos J, Jourdan M, et al. Cooperation between heparin-binding EGF-like growth factor and interleukin-6 in promoting the growth of human myeloma cells. Oncogene 2002; 21: 2584-92. ArticlePubMedPDF
- 32. Sugiura T, Berditchevski F. Function of alpha3beta1-tetraspanin protein complexes in tumor cell invasion: evidence for the role of the complexes in production of matrix metalloproteinase 2 (MMP-2). J Cell Biol 1999; 146: 1375-89. PubMedPMC
REFERENCES
Figure & Data
References
Citations

- Predicting patient outcomes with gene-expression biomarkers from colorectal cancer organoids and cell lines
Alexandra Razumovskaya, Mariia Silkina, Andrey Poloznikov, Timur Kulagin, Maria Raigorodskaya, Nina Gorban, Anna Kudryavtseva, Maria Fedorova, Boris Alekseev, Alexander Tonevitsky, Sergey Nikulin
Frontiers in Molecular Biosciences.2025;[Epub] CrossRef - Intestinal estrogen receptor beta modulates the murine colon tumor immune microenvironment
Madeleine Birgersson, Matilda Holm, Carlos J. Gallardo-Dodd, Baizhen Chen, Lina Stepanauskaitė, Linnea Hases, Claudia Kutter, Amena Archer, Cecilia Williams
Cancer Letters.2025; 622: 217661. CrossRef - Establishment of Coala: a novel 3D and 2D cancer cell line derived from colorectal cancer liver metastasis
Sandra Mersakova, Juraj Marcinek, Dusan Brany, Olga Chodelkova, Martin Vorcak, Martin Cermak, Martina Poturnajova, Zuzana Kozovska, Henrieta Skovierova, Slavomira Novakova, Barbora Mitruskova, Dusan Loderer, Marian Grendar, Veronika Holubekova, Andrea Hor
Human Cell.2025;[Epub] CrossRef - CD9, a tetraspanin in cancer: biology and therapeutic promise in acute leukemia
Océane Guého, Elie Cousin, Jérémie Rouger-Gaudichon, Anne-Gaëlle Rio, Sébastien Corre, Virginie Gandemer, Frédéric Mazurier
Oncogenesis.2025;[Epub] CrossRef - Proteomic analysis of plasma exosomes in patients with metastatic colorectal cancer
Zhaoyue Zhong, Jiayin Ji, Hongxia Li, Ling Kang, Haipeng Zhu
Clinical Proteomics.2024;[Epub] CrossRef - Prognostic value and multifaceted roles of tetraspanin CD9 in cancer
Róbert Ondruššek, Barbora Kvokačková, Karolína Kryštofová, Světlana Brychtová, Karel Souček, Jan Bouchal
Frontiers in Oncology.2023;[Epub] CrossRef - Proteomic Signature of Extracellular Vesicles Associated with Colorectal Cancer
Natalia Soloveva, Svetlana Novikova, Tatiana Farafonova, Olga Tikhonova, Victor Zgoda
Molecules.2023; 28(10): 4227. CrossRef - Anti-Human CD9 Fab Fragment Antibody Blocks the Extracellular Vesicle-Mediated Increase in Malignancy of Colon Cancer Cells
Mark F. Santos, Germana Rappa, Simona Fontana, Jana Karbanová, Feryal Aalam, Derek Tai, Zhiyin Li, Marzia Pucci, Riccardo Alessandro, Chikao Morimoto, Denis Corbeil, Aurelio Lorico
Cells.2022; 11(16): 2474. CrossRef - The Study of the Extracellular Matrix in Chronic Inflammation: A Way to Prevent Cancer Initiation?
Asia Marangio, Andrea Biccari, Edoardo D’Angelo, Francesca Sensi, Gaya Spolverato, Salvatore Pucciarelli, Marco Agostini
Cancers.2022; 14(23): 5903. CrossRef - In vivo expansion of a CD9+ decidual-like NK cell subset following autologous hematopoietic stem cell transplantation
Ane Orrantia, Enrique Vázquez-De Luis, Gabirel Astarloa-Pando, Iñigo Terrén, Ainhoa Amarilla-Irusta, Diego Polanco-Alonso, Carmen González, Alasne Uranga, Tomás Carrascosa, Juan J. Mateos-Mazón, Juan C. García-Ruiz, Sergio Callejas, Ana Quintas, Ana Dopaz
iScience.2022; 25(10): 105235. CrossRef - Inhibition of cancer-cell migration by tetraspanin CD9-binding peptide
Thanawat Suwatthanarak, Masayoshi Tanaka, Yoshitaka Miyamoto, Kenji Miyado, Mina Okochi
Chemical Communications.2021; 57(40): 4906. CrossRef - High expression of tumor susceptibility gene 101 (TSG101) is associated with more aggressive behavior in colorectal carcinoma
Elmira Gheytanchi, Leili Saeednejad Zanjani, Roya Ghods, Maryam Abolhasani, Marzieh Shahin, Somayeh Vafaei, Marzieh Naseri, Fahimeh Fattahi, Zahra Madjd
Journal of Cancer Research and Clinical Oncology.2021; 147(6): 1631. CrossRef - Increased CD9 expression predicts favorable prognosis in human cancers: a systematic review and meta-analysis
Hyun Min Koh, Bo Gun Jang, Dong Hui Lee, Chang Lim Hyun
Cancer Cell International.2021;[Epub] CrossRef - Prognostic Value of CD9 in Solid Tumor: A Systematic Review and Meta-Analysis
Ping Zeng, Meng Si, Rui-xia Sun, Xu Cheng, Xiao-yang Li, Min-bin Chen
Frontiers in Oncology.2021;[Epub] CrossRef - Matrix Effect in the Isolation of Breast Cancer-Derived Nanovesicles by Immunomagnetic Separation and Electrochemical Immunosensing—A Comparative Study
Silio Lima Moura, Mercè Martì, María Isabel Pividori
Sensors.2020; 20(4): 965. CrossRef - CD9 expression indicates a poor outcome in acute lymphoblastic leukemia
Peiqi Liang, Miao Miao, Zhuogang Liu, Hongtao Wang, Wei Jiang, Shiyu Ma, Chuan Li, Rong Hu
Cancer Biomarkers.2018; 21(4): 781. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure




Fig. 1.
Fig. 2.
Fig. 3.
Fig. 4.
| Variable | CD9 expression in tumor cells (T-CD9) |
CD9 expression in immune cells (I-CD9) |
||||||
|---|---|---|---|---|---|---|---|---|
| No. of cases | Low | High | p-value | No. of cases | Low | High | p-value | |
| Age (yr) | .449 | .496 | ||||||
| ≤ 60 | 204 | 83 (64.3) | 121 (69.1) | 205 | 25 (62.5) | 180 (67.9) | ||
| > 60 | 100 | 46 (35.7) | 54 (30.9) | 100 | 15 (37.5) | 85 (32.1) | ||
| Sex | .175 | .581 | ||||||
| Male | 163 | 75 (58.1) | 88 (50.3) | 163 | 23 (57.5) | 140 (52.8) | ||
| Female | 141 | 54 (41.9) | 87 (49.7) | 142 | 17 (42.5) | 125 (47.2) | ||
| Tumor location | .224 | .385 | ||||||
| Right | 89 | 33 (25.6) | 56 (32.0) | 89 | 14 (35.0) | 75 (28.3) | ||
| Left | 215 | 96 (74.4) | 119 (68.0) | 216 | 26 (65.0) | 190 (71.7) | ||
| Gross type |
.775 | .632 | ||||||
| Fungating | 137 | 61 (48.0) | 76 (46.3) | 137 | 16 (43.2) | 121 (47.5) | ||
| Infiltrative | 154 | 66 (52.0) | 88 (53.7) | 155 | 21 (56.8) | 134 (52.5) | ||
| AJCC TNM stage | .938 | .156 | ||||||
| Stage I | 32 | 13 (10.1) | 19 (10.8) | 33 | 1 (2.5) | 32 (12.1) | ||
| Stage II | 134 | 56 (43.4) | 78 (44.6) | 134 | 21 (52.5) | 113 (42.6) | ||
| Stage III | 138 | 60 (46.5) | 78 (44.6) | 138 | 18 (45.0) | 120 (45.3) | ||
| Tumor depth (pT) | .909 | .137 | ||||||
| pT1 | 9 | 4 (3.1) | 5 (2.8) | 9 | 0 | 9 (3.4) | ||
| pT2 | 30 | 11 (8.5) | 19 (10.9) | 31 | 1 (2.5) | 30 (11.3) | ||
| pT3 | 233 | 101 (78.3) | 132 (75.4) | 233 | 36 (90.0) | 197 (74.3) | ||
| pT4 | 32 | 13 (10.1) | 19 (10.9) | 32 | 3 (7.5) | 29 (11.0) | ||
| LN metastasis (pN) | .884 | .935 | ||||||
| pN0 | 167 | 69 (53.5) | 98 (56.0) | 168 | 22 (55.0) | 146 (55.1) | ||
| pN1 | 86 | 37 (28.7) | 49 (28.0) | 86 | 12 (30.0) | 74 (27.9) | ||
| pN2 | 51 | 23 (17.8) | 28 (16.0) | 51 | 6 (15.0) | 45 (17.0) | ||
| Tumor differentiation |
.734 | .014 | ||||||
| WD/MD | 261 | 110 (85.9) | 151 (87.3) | 262 | 29 (74.4) | 233 (88.6) | ||
| PD/Other | 40 | 18 (14.1) | 22 (12.7) | 40 | 10 (25.6) | 30 (11.4) | ||
| Lymphatic invasion |
.394 | .361 | ||||||
| Absent | 192 | 77 (67.5) | 115 (72.3) | 193 | 21 (63.6) | 172 (71.4) | ||
| Present | 81 | 37 (32.5) | 44 (27.7) | 81 | 12 (36.4) | 69 (28.6) | ||
| Venous invasion |
.467 | .300 | ||||||
| Absent | 208 | 85 (74.6) | 123 (78.3) | 209 | 23 (69.7) | 186 (77.8) | ||
| Present | 63 | 29 (25.4) | 34 (21.7) | 63 | 10 (30.3) | 53 (22.2) | ||
| Perineural invasion |
.553 | .856 | ||||||
| Absent | 195 | 84 (75.0) | 111 (78.2) | 196 | 25 (78.1) | 171 (76.7) | ||
| Present | 59 | 28 (25.0) | 31 (21.8) | 59 | 7 (21.9) | 52 (23.3) | ||
| Parameter | Univariate analysis |
p-value | Multivariate analysis I |
p-value | Multivariate analysis II |
p-value |
|---|---|---|---|---|---|---|
| Hazard ratio (95% confidence interval, mo) | Hazard ratio (95% confidence interval, mo) | Hazard ratio (95% confidence interval, mo) | ||||
| AJCC stage | <.001 | .002 | .001 | |||
| Stage l/ll | Reference | Reference | Reference | |||
| Stage III | 3.831 (2.210–6.644) | 2.727 (1.448–5.136) | 2.807 (1.495–5.272) | |||
| Lymphatic invasion | <.001 | .121 | .101 | |||
| Absent | Reference | Reference | Reference | |||
| Present | 2.852 (1.647-4.938) | 1.794 (0.858–3.755) | 1.843 (0.887–3.831) | |||
| Vascular invasion | .001 | .461 | .485 | |||
| Absent | Reference | Reference | Reference | |||
| Present | 2.552 (1.438-4.529) | 1.330 (0.623–2.838) | 1.308 (0.616–2.777) | |||
| Perineural invasion | .006 | .163 | .143 | |||
| Absent | Reference | Reference | Reference | |||
| Present | 2.202 (1.248–3.887) | 1.540 (0.840–2.824) | 1.577 (0.857–2.905) | |||
| T-CD9 | .023 | .167 | Not included | |||
| Low | Reference | Reference | - | |||
| High | 0.558 (0.337–0.922) | 0.669 (0.379–1.183) | - | |||
| T-CD9/I-CD9 | .043 | Not included | .199 | |||
| T-CD9 (+)/l-CD9 (–) | Reference | - | Reference | |||
| T-CD9 (+)/l-CD9 (+) and T-CD9 (–)/l-CD9 (–) | 3.591 (0.489–26.351) | .209 | - | - | 3.136 (0.419–23.459) | .266 |
| T-CD9 (–)/l-CD9 (+) | 5.983 (0.816–43.860) | .078 | - | - | 4.492 (0.600–33.611) | .143 |
Values are presented as number (%). AJCC, American Joint Committee on Cancer; LN, lymph node; WD, well differentiated; MD, moderately differentiated; PD, poorly differentiated. One case is excluded from this statistical analysis due to unavailable TMA data; Only patients with available clinicopathological data are included.
CRC, colorectal carcinoma; AJCC, American Joint Committee on Cancer; T-CD9, CD9 expression in tumor cells; I-CD9, CD9 expression in immune cells.

 E-submission
E-submission