Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 54(6); 2020 > Article
-
Original Article
The frequency of POLE-mutation in endometrial carcinoma and prognostic implications: a systemic review and meta-analysis -
Alaa Salah Jumaah1
 , Mais Muhammed Salim1
, Mais Muhammed Salim1 , Hawraa Sahib Al-Haddad2
, Hawraa Sahib Al-Haddad2 , Katherine Ann McAllister3
, Katherine Ann McAllister3 , Akeel Abed Yasseen1
, Akeel Abed Yasseen1
-
Journal of Pathology and Translational Medicine 2020;54(6):471-479.
DOI: https://doi.org/10.4132/jptm.2020.07.23
Published online: September 2, 2020
1Department of Pathology and Forensic Medicine, Faculty of Medicine, University of Kufa, Kufa, Iraq
2Al-Furat Al-Awsat Hospital, Kufa, Iraq
3School of Biomedical Science, University of Ulster, Northern Ireland, UK
- Corresponding Author: Akeel Abed Yasseen, PhD, Department of Pathology and Forensic Medicine, Faculty of Medicine, University of Kufa, Kufa, P.O. Box 21, Najaf Governorate, Iraq Tel: +96-47811131586, E-mail: akeelyasseen@uokufa.edu.iq
© 2020 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- Endometrial carcinoma (EC) is classified into four distinct molecular subgroups including ultramutated DNA polymerase epsilon (POLE). POLE-mutated tumors have the best prognosis and are a promising target for immunotherapy. This meta-analysis consolidated the reported variation of POLE-mutant frequency and assessed prognostic value in EC.
-
Methods
- Internet searches explored scientific data bases: EMBASE, PubMed, and the Cochrane Central Register of Controlled Trials databases. Data was extracted from eligible studies including: sample size, number of positive POLE-mutant cases, sequencing information, clinicopathologic data, and survival data. Meta-analysis and a random-effects model produced pooled estimates of POLE frequency and prognostic parameters using 95% confidence intervals (CI), hazard ratios (HR), and odd ratios (OR).
-
Results
- Six thousand three hundred and forty-six EC patient cases were pooled from 25 studies. The pooled proportion of POLE gene mutation in EC was 8.59% (95% CI, 7.01 to 10.32), of which 8.22% (95% CI, 6.27 to 10.42) were type I and 0.93% (95% CI, 0.34 to 1.81) type 2. Clinicopathologic data showed that POLE-mutated tumors are mostly endometrioid. They present at higher levels in earlier stages (I–II) of EC (89.51%; 95% CI, 81.11 to 95.66) at the highest grade III (51.53%; 95% CI, 36.08 to 66.84) with reduced myometrial invasion (OR, 1.48, 95% CI, 0.99 to 2.20). Survival analysis indicated favorable overall survival (HR, 0.90), disease-specific survival (HR, 0.41), and progression-free survival (HR, 0.23) for POLE mutant EC.
-
Conclusions
- Almost one-tenth of EC patients have POLE-mutated tumors. Given their improved prognostic potential, identifying the POLE mutation status is key for the management of EC patients.
- This study was conducted according to the guidelines of Preferred Reporting for Systemic Review and Meta-analysis (PRISMA) statement [32].
- Literature search strategy
- Searches were conducted according to the guidelines of PRISMA statement 2009 [32]. Two authors (A.S.J. and H.S.A.) searched independently the following data bases from inceptions to October 2019: Embase, PubMed, Cochrane central Register of Controlled trials, and Ovid. The reference lists were also scanned within the articles. There were no language limits and international papers were translated. All pathology and oncology journals indexed in the Scimago directory were reviewed and relevant papers scanned (A.S.J. and M.M.S.). The following search terms were used:
- 1) ‘Endometrial cancer’ or ‘uterine cancer.’
- 2) ‘POLE gene’ or ‘ultramutated endometrial carcinoma.’
- Inclusion criteria
- The following patient inclusion criteria were used:
- 1) EC or one of its histological variants was present in patients.
- 2) The expression of POLE gene was reported using genetic
- testing (e.g. sequencing, Sanger sequencing, next generation sequencing, polymerase chain reaction).
- 3) A full paper was published and studies published in abstract format only were excluded.
- 4) When similar studies were generated from the same patient, only the most recent investigation was included.
- Study selection
- Studies were identified using different data bases. The title of the paper and abstract were assessed by two independent authors (A.S.J. and H.S.A.). The full texts were also reviewed independently by two authors (M.M.S. and K.A.M.). Any disagreement was resolved under guidance of the senior author (A.A.Y.).
- Data collection
- The following data was extracted from eligible studies by two authors (A.S.J. and H.S.A.): study information (first author and year of publication), patient characteristics (sample size and gender), site of the study, and test method for POLE gene and proportion detected. Clinicopathologic data extracted included tumor stage and grade, presence of lymphovascular and myometrial invasion, and patient survival. If the relevant data was not available, it was recorded as NR (not reported). All datasets were checked independently (M.M.S.). Any disagreements were resolved by discussion and consultation with the senior author (A.A.Y.).
- Meta-analysis and statistical methods
- The proportion of endometrial cancers that harbor POLE gene mutations was calculated using medcalc software [33]. The pooled proportion of POLE was calculated using the random effect model [32] for meta-analysis. For clinicopathologic meta-analysis, proportions of tumour stage, grade, lymphovascular invasion, myometrial invasion (MI), and survival analysis (overall survival, disease-free survival, and progression-free survival) were pooled from each study. The variation between datasets was assessed using the heterogeneity test with inconsistency index (I2) and Q statistic. The level of study heterogeneity was considered low at 25% (I2=25%), medium at 50% (I2=50%) or high at 75% (I2= 75%). In regard to the Q statistic, a p-value of less than 0.1 was considered to represent significant heterogeneity. The possibility of publication bias was assessed by visual method using a funnel plot. This determined funnel plot asymmetry resulting from factors such as non-publication of studies with negative results.
- Sensitivity and subgroup analysis
- Lastly, sensitivity analysis was conducted by omitting each study one-by-one to discover its contribution on the pooled meta-analysis results. Subgroup analysis was performed according to geographical area (Asia, West-Europe, and America) and to different histological types to discover sources of heterogeneity. The subgroup analysis was further extended by using a meta regression model.
MATERIALS AND METHODS
- Study selection for meta-analysis
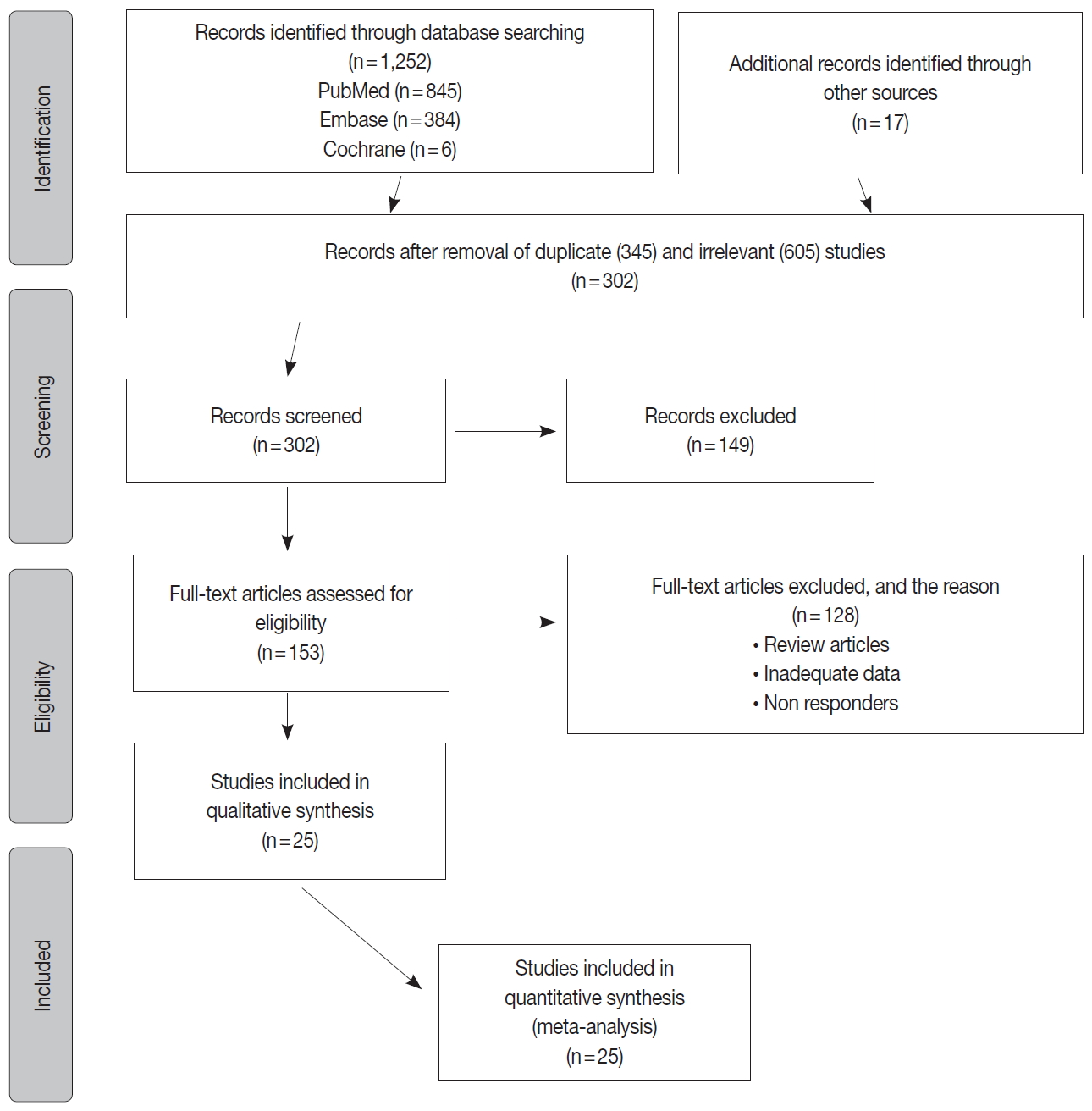
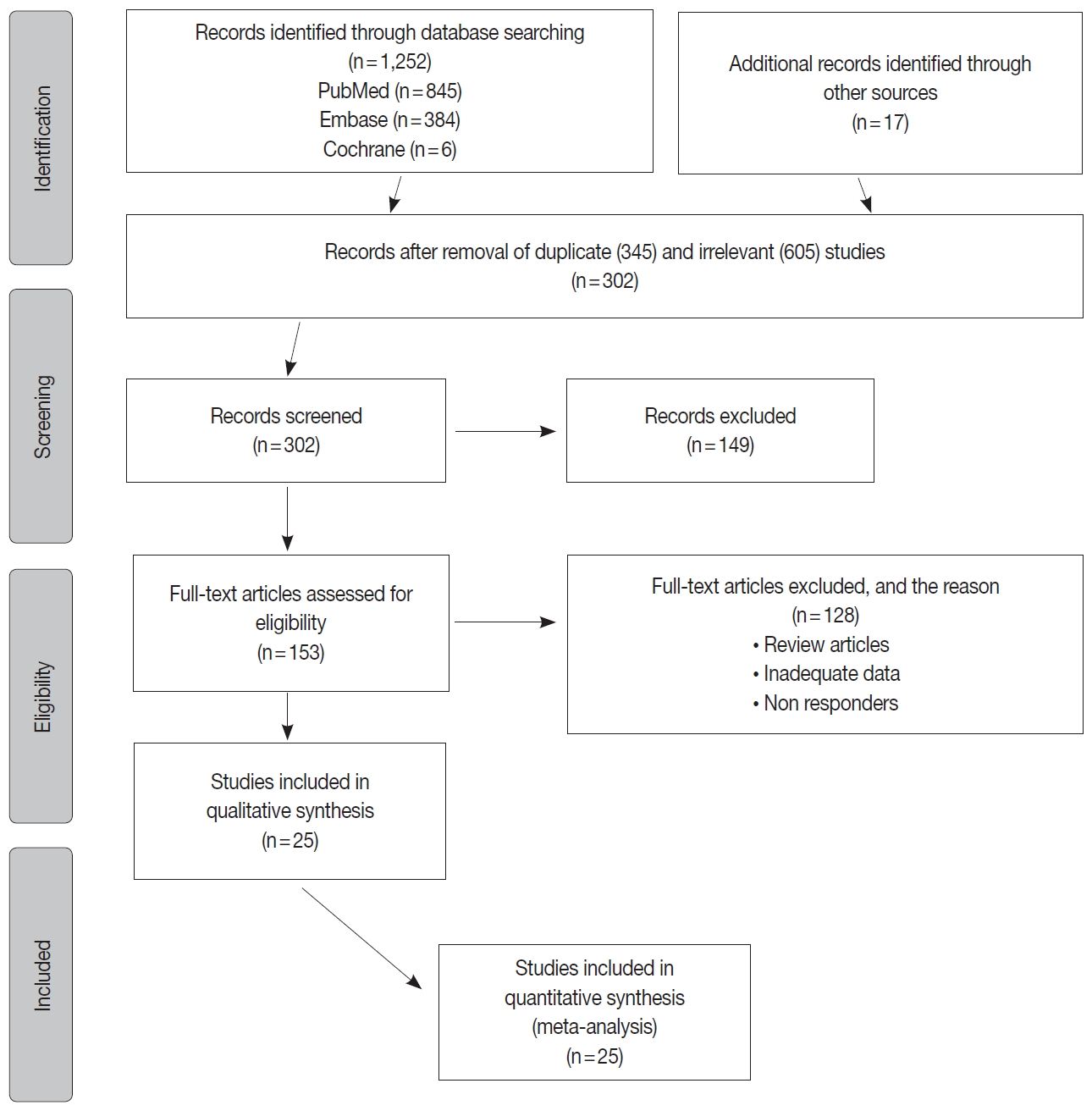
- The inclusion criteria were met by 1,252 studies, of which 345 studies were excluded for duplication, and 605 studies were excluded as irrelevant by reviewing the title and abstract. The full texts of the remaining 302 studies were considered for review. Approximately 25 studies were left for the final meta-analysis as illustrated in the flowchart diagram (Fig. 1). All of the studies were published from 2013–2019, and there were no unpublished studies of relevance. The study characteristics and POLE gene sequencing methods are listed in (Table 1). The majority used Sanger sequencing to detect tumor mutations in exons 9–14 of the POLE exonuclease domain (Table 1) [8-29].
- Meta-analysis of EC
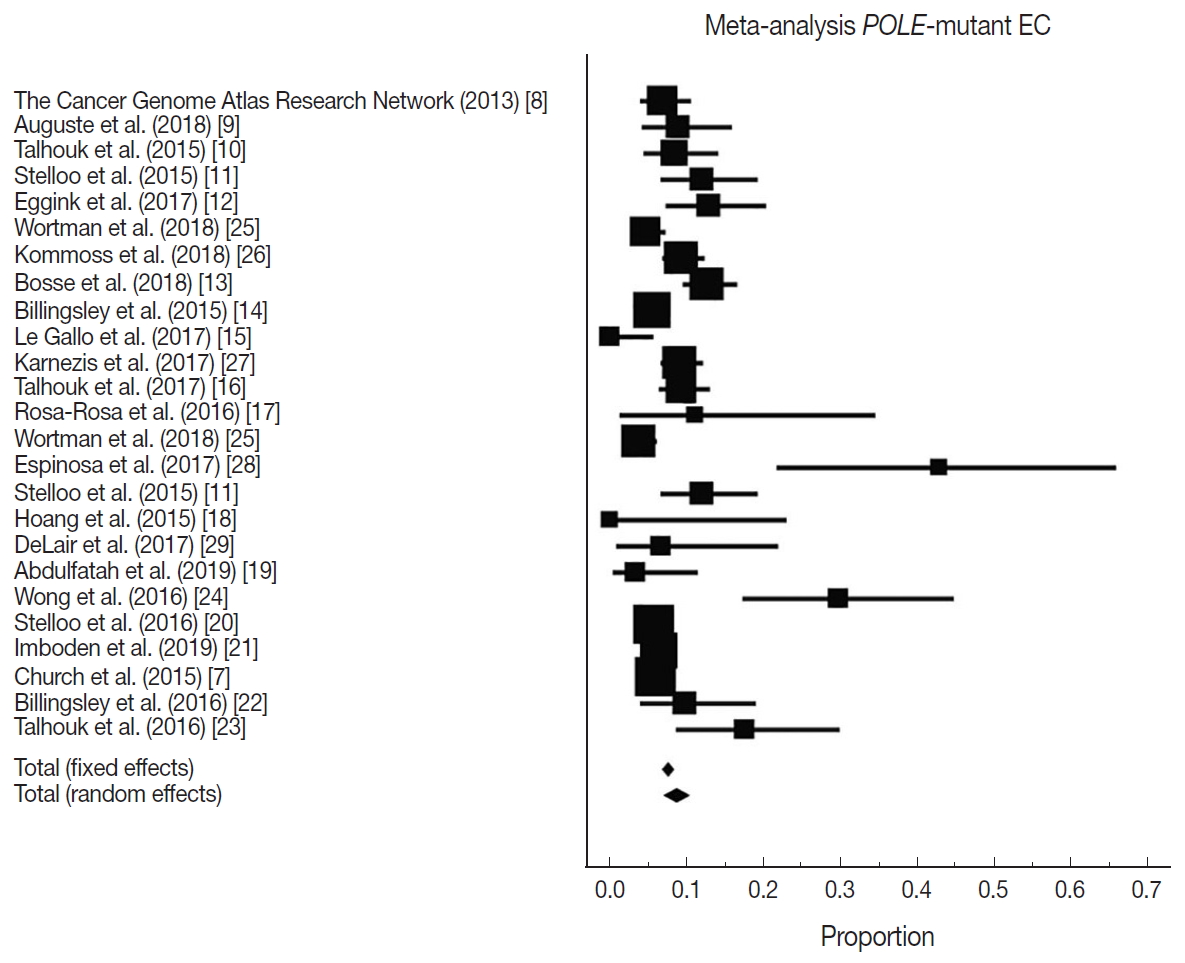
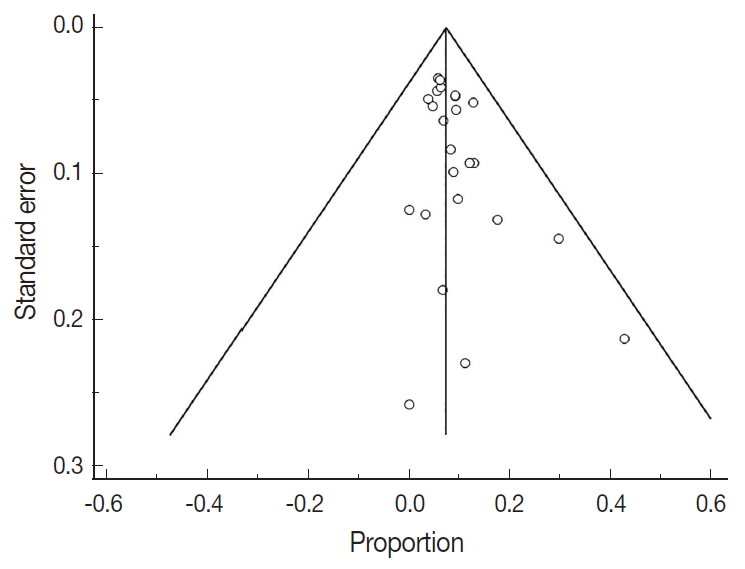
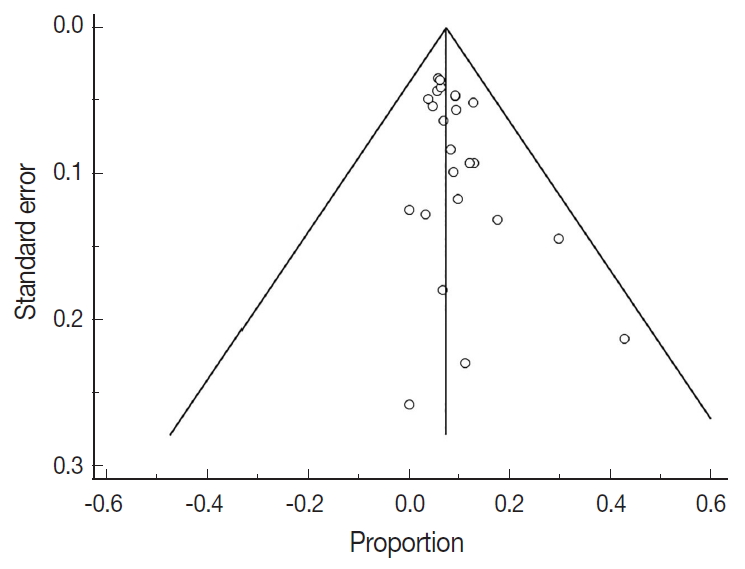
- There were 6,346 EC patient cases pooled from the 25 studies and investigated for POLE gene tumor alterations. All studies (except one) were performed in western countries. The studies were published in English and full texts obtained. There was significant heterogeneity between the studies (Q=109.57, I2 =78.10%; 95% confidence interval [CI] for I2, 68.15 to 84.94; p<.001). The random effect model was used for final meta-analysis because the significant heterogeneity. The pooled proportion of POLE gene mutated in EC was determined at 8.59% (95% CI, 7.01 to 10.32) as shown in Table 2. A forest plot representation of the EC patient cases comprising each study included for meta-analysis is shown in Fig. 2. Publication bias was assessed by funnel plot (Fig. 3) and the visual assessment was symmetrical.
- Analysis of EC histologic variants type I and type II
- There were 3,363 patients included for the type 1 meta-analysis using a total of 9 studies (Supplementary Fig. S1). The mutated POLE gene proportion in type I EC was 8.22% (95% CI, 7.01 to 10.32). There was significant heterogeneity between studies (I2 inconsistency=74.88%; 95% CI, 51.43 to 87.00; p<.001) as shown in Table 2. A total of 3,423 patients with EC type II were obtained from 10 studies for the final meta-analysis (Supplementary Fig. S2). The histological types in EC type II are show in (Supplementary Table S1). The pooled proportion of mutated POLE gene in type II was 0.93% (95% CI, 0.34 to 1.81) with a high I2 value (75.32%; 95% CI, 54.08 to 86.74; p<.001).
- Subgroup analysis (country of origin)
- In order to explore the role of heterogeneity, a subgroup analysis was performed according to the study site (Supplementary Fig. S3, S4). The studies were separated into two groups according to geographical area (USA and Canada vs. European countries). The heterogeneity was higher (I2, inconsistency) for POLE genetic analysis in the European countries (78.41%; 95% CI, 59.27 to 88.55) compared to the USA and Canada (37.41%; 95% CI, 0.00 to 69.22).
- Sensitivity analysis
- There were 2 identified study outliers [24,28]. Meta-analysis was performed after outlier exclusion. There was still significant heterogeneity (I2=70.39% [95% CI for I2, 54.78 to 80.61; p<.001]) and exclusion did not significantly affect the final pooled proportion (pooled proportion=7.76 [95% CI, 6.45 to 9.18]) (Supplementary Fig. S5). There were also two studies without POLE gene mutated EC [15,18]. Here, the pooled proportion was re-calculated using the random effect model (Supplementary Fig. S6). Significant heterogeneity was still present (I2= 76.31%; 95% CI for I2, 64.98 to 83.97; p<.001). There were 10 studies [9,13,15,17,19-21,23,24,30] identified with a low sample size (<100). These cases were excluded and the pooled proportions re-estimated (Supplementary Fig. S7). There was still significant heterogeneity (I2=74.41%; 95% CI for I2, 53.56 to 85.90; p<.001).
- Meta-regression
- Meta-regression was performed to identify the source of heterogeneity using the study country of origin and the POLE gene detection method (Supplementary Table S2). There was increased heterogeneity when considering the site of study performance in the European countries (I2=78.41%) compared to United States and Canadian studies (I2=37.41%).
- Clinicopathological characteristics
- Clinicopathologic data was extracted from eligible studies of POLE mutant EC for meta-analysis (Supplementary Fig. S8-S14). The pooled proportions for stage, grade, lymphovascular invasion (LVI) and MI parameter are reported in Table 2. High heterogeneity was noted for pooled stage, grade, and LVI data. The pooled odd ratios were also calculated for POLE-mutant versus wild type POLE according to each clinicopathologic variable (Table 3).
- Tumor stage and grade in POLE mutant EC
- The pooled proportion of mutant POLE presented at high levels of 89.51% (95% CI, 81.11 to 95.66) at the earliest EC stages of I–II (Table 2). This reduced to 14.77% by stages III–IV (95% CI, 5.99 to 26.59). The pooled odd ratio of stage I–II POLE mutant EC versus wild type was 3.72 (2.06 to 6.73), while stage III–IV was 0.26 (95% CI, 0.14 to 0.49) (Table 2, Supplementary Fig. S15, S16). The pooled collective proportions of grade I–II POLE mutant tumors was lower at 46.36% (95% CI, 30.66 to 62.43) compared to 51.53 of grade III (95% CI, 36.08 to 66.844) as shown in Table 2. The pooled odd ratio of grade I–II POLE mutant EC versus wild type tumors was 0.40 (95% CI, 0.29 to 0.54) (Supplementary Fig. S17) with low heterogeneity (I2=3.95%, 95% CI, 0.00 to 69.18). Therefore, POLE mutated tumors present with a higher grade but at lower stage than wild type POLE mutant EC.
- LVI and MI in POLE-mutant EC
- The pooled proportion of LVI was 31.11% (95% CI, 10.44 to 56.86). The pooled odd ratio of LVI positive in POLE mutant EC vs. wild type EC was 0.92 (0.643 to 1.34) (Table 3). The pooled proportion of MI either less or greater than 50% of myometrium in POLE-mutant tumor was equal at 49.90% (95% CI, 43.71 to 56.21) and 49.05% (95% CI, 39.17 to 58.98), respectively. Heterogeneity was low for pooled MI data (<50% myometrium) at 22.10%; 95% CI, 0.00 to 65.16 (Table 2, Supplementary Figs. S13, S14). The pooled odd ratio of MI <50% in POLE mutant EC versus MI < 50% in wild type POLE EC was 1.481 (95% CI, 0.99 to 2.20) with 47.63% heterogeneity (95% CI, 0.00 to 79.24) (Table 3). Overall, these findings imply that POLE mutant EC tumors have reduced ability to progress to myometrial invasion, which is an important prognostic finding.
- Endometrioid and non-endometrioid histologic types in POLE mutant EC
- POLE mutant tumors were found to mainly present with endometrioid histologic type. The pooled proportions of type I and type II EC are shown in Table 2. The pooled odd ratio of endometrioid (type I) POLE mutant EC vs. endometrioid (type I) in wild type POLE EC was 1.72 (95% CI: 1.11 to 2.66), with very low heterogeneity (I2=0.00%; 95% CI: 0.00 to 68.45) (Table 3, Supplementary Fig. S18).
- Survival analysis
- The studies used for survival meta-analysis are listed in Table 4. Survival analysis was expressed using overall survival (OS), disease-specific survival (DSS), and progression-free survival (Supplementary Fig. S19–S21). All the survival parameters had a hazard ratio (HR) of less than 1. The estimated HR for OS was 0.90 (95% CI, 0.59 to 1.38) with low heterogeneity 0.00% (95% CI, 0.00 to 73.28). On the other hand, estimated HR for DSS was 0.41 (95% CI, 0.30 to 0.55), also with low heterogeneity (0.00%; 95% CI, 0.00 to 67.34). Likewise, the estimated HR of progression-free survival in POLE-mutant EC was 0.23 (95% CI, 0.08 to 0.64) with a low level of heterogeneity (I2=0.00%, 95% CI, 0.00 to 0.00). These findings indicate better survival and favorable prognosis in POLE mutant EC patients.
RESULTS
- EC is the most common gynecologic malignancy in the western world [2,3,34], and survival rates are not improving. There is urgent need for strategies to improve outlook for patients with aggressive subtypes and advanced disease. TCGA first identified the very interesting molecular subset of POLE-ultramutated EC [1] that features a favorable prognostic potential, despite high tumor grading. Many follow-up studies investigated POLE mutant EC tumors, however the frequency reported is variable [8-31]. Some studies show ultra-high levels of mutation at 42.9% [28] compared to zero in others [15,18]. This meta-analysis aimed to resolve these datasets to estimate POLE gene mutational frequency in EC and the overall effect on patient prognosis.
- Our meta-analysis determined that 8.595% of endometrial tumors harbor POLE gene mutations. The majority have endometrioid histology and present at the earlier stage of disease progression. Paradoxically the POLE mutant tumors present at he highest grade (grade III, 51.5%) and yet have a better outcome with survival analysis. They also have reduced ability to progress to myometrial invasion which is an important prognostic marker. Many studies confirm that POLE mutant tumors have a better prognosis [6-8,13,22,28]. POLE mutations in high grade (grade III) endometrioid EC are shown to be associated with a lower risk of recurrence and death [22]. The presence of POLE mutations even offers a favorable prognosis for rare and aggressive undifferentiated EC [28]. POLE mutation could potentially act as a prognostic biomarker to guide treatment of women with grade III, early-stage disease, with either the common endometrioid or more rare histological types. The lower risk of occurrence means that administration of adjuvant therapy for these patient subsets could be inappropriate. POLE proofreading mutations have also been shown to elicit an anti-tumor response [35]. There is now an emerging link between high mutation burden in tumors, the immune response and improved prognosis in cancer patients. Indeed, ultramutated POLE tumors have been shown to feature higher immune infiltrations and programmed death-1 and programmed death-ligand 1 expression [36]. These immune cells may offset the survival risk caused by higher tumor grades in ultramutated POLE. Overall, mutant POLE is a key proportion of EC patients to target therapeutically for maximizing clinical impact, and a future target for immunotherapy [9].
- However, it is important to note that significant heterogeneity was present in this meta-analysis. Indeed, heterogeneity is a key problem for meta-analysis studies. We tried to resolve the heterogeneity sources. Initially, all studies were re-checked with respect to data extraction and entry. The sensitivity analysis was conducted by re-estimating the pooled proportion of POLE gene after exclusion of outlier studies and studies with low sample size. We also adopted a random effect model for final pooled estimation as this model assumed that effects estimated in different studies are not identical. We also tried to perform subgroup analysis with respect to patient age but unfortunately this parameter was not recorded in the majority of studies. Publication bias was investigated; in our study the plot was relatively symmetrical, indicating that publication bias is unlikely [37]. The presence of high heterogeneity was reduced by subgroup and meta-regression according to geographical distribution. European studies were found to be the main contributor to the heterogeneity. Therefore, data obtained from different countries can cause confounding effects and is a potential study limitation. The issue of heterogeneity in genetic studies is also further compounded with recent advances in sequencing technology. The improved yield of genetic testing can increase detection of variants of unknown significance. However in our POLE gene study the lack of standardized clarification of variants of unknown significance may have contributed to heterogeneity. This will be a key area to investigate during future studies.
DISCUSSION
- Our meta-analysis consolidates previous study estimates of POLE-mutated EC frequency and confirms its prognostic benefit for patients. The status and frequencies of the POLE gene mutation in EC has implications for medical management and future administration of immunotherapy. The POLE mutational status serves as an important prognostic marker, and grade III, early-stage disease patients with endometrioid histology could favor a change in medical management.
Conclusion
Supplementary Materials
Ethics Statement
The present study was approved by the Institutional Review Board of the University of Kufa (IRB approval No. UK-2018-0456) in accordance with the 1964 Helsinki declaration and its later amendments. Formal written informed consent was not required with a waiver issued by the Institutional Review Board of the University of Kufa. All the authors will be held responsible for any false statements or failure to follow the ethical guidelines.
Author contributions
Conceptualization: ASJ, MMS. Data curation: ASJ. Formal analysis: ASJ, MMS. Methodology: ASJ, HSA, MMS. Project administration: HSA, MMS. Resources: HSA. Software: ASJ, MMS. Supervision: ASJ, AAY. Validation: MMS, HSA. Visualization: HSA, MMS. Writing—original draft: ASJ, AAY, KAM. Writing—review & editing: AAY, ASJ, KAM. Approval of final manuscript: all authors.
Conflicts of Interest
The authors declare that they have no potential conflicts of interest.
Funding Statement
No funding to declare.

| Study | EC cohort size | Proportion POLE-mutant | Country | Sequencing method | Location of exonuclease mutations |
|---|---|---|---|---|---|
| The Cancer Genome Atlas Research Network (2013) [8] | 248 | 17 | USA | Exome sequencing | Hotspots: Pro286Arg and Val411Leu |
| Auguste et al. (2018) [9] | 102 | 9 | Canada and Europe | Sanger sequencing | Exons 9 and 13 |
| Talhouk et al. (2015) [10] | 143 | 12 | Canada | Fluidigm-MiSeq and sanger sequencing | Exons 9–14 |
| Stelloo et al. (2015) [11] | 116 | 14 | Europe and Australia | Sanger sequencing | Exons 9 and 13 |
| Eggink et al. (2017) [12] | 116 | 15 | Europe and Australia | Sanger sequencing | Exons 9, 13 and 14 |
| Wortman et al. (2018) [25] | 344 | 16 | Netherlands | Sequencing | Not reported |
| Kommoss et al. (2018) [26] | 452 | 42 | Germany | Sequencing | Exons 9–14 |
| Bosse et al. (2018) [13] | 376 | 48 | USA, Canada, and Europe | Sanger or next-generation approaches | Hotspots in the exonuclease domain (exons 9–14) |
| Billingsley et al. (2015) [14] | 535 | 30 | USA | Sanger sequencing | Residues 268–471 |
| Le Gallo et al. (2017) [15] | 63 | 0 | USA and Europe | Sanger sequencing | Not reported |
| Karnezis et al. (2017) [27] | 460 | 42 | Canada | Sequencing | Not reported |
| Talhouk et al. (2017) [16] | 319 | 30 | Canada | Sanger sequencing | Exons 9–14 |
| Rosa-Rosa et al. (2016) [17] | 18 | 2 | USA and Europe | Sanger sequencing | Exons 9 and 13 |
| Wortman et al. (2018) [25] | 416 | 16 | Netherlands | Sequencing | Not reported |
| Espinosa et al. (2017) [28] | 21 | 9 | Spain | Sequencing | Exons 9 to 14 |
| Stelloo et al. (2015) [11] | 116 | 14 | Europe | Sanger sequencing | Exons 9 and 13 |
| Hoang et al. (2015) [18] | 14 | 0 | Canada | Sanger sequencing | Exons 9–14 |
| DeLair et al. (2017) [29] | 30 | 2 | USA | Sequencing | Exons 9–14 |
| Abdulfatah et al. (2019) [19] | 60 | 2 | USA | Sanger sequencing | Exons 9 and 13 |
| Wong et al. (2016) [24] | 47 | 14 | Singapore | Next generation sequencing | Exons 9–14 |
| Stelloo et al. (2016) [20] | 834 | 49 | Netherlands | Sanger sequencing | Exons 9 and 13 |
| Imboden et al. (2019) [21] | 599 | 38 | Sweden | Sanger sequencing | Exons 9–14 |
| Church et al. (2015) [7] | 788 | 48 | Europe | Sanger sequencing | Exons 9 and 13 |
| Billingsley et al. (2016) [22] | 72 | 7 | USA | Sanger sequencing | Residues 268–471 |
| Talhouk et al. (2016) [23] | 57 | 10 | USA and Canada | Ultra-deep MiSeq or sanger sequencing | Exons 9–14 |
| Study | OS estimated HR (95% CI) | DSS estimated HR (95% CI) | PFS estimated HR (95% CI) | Survival analysis test | Method |
|---|---|---|---|---|---|
| Talhouk et al. (2017) [16] | 1.01 (0.29–3.42) | 0.42 (0.30–0.57) | - | Multivariable analysis | Kaplan-Meier survival analysis |
| Talhouk et al. (2015) [10] | 0.17 (0.01–1.98) | 0.170 (0.01–1.99) | - | Multivariable analysis | Kaplan-Meier with log-rank significance testing and Cox proportional hazard regression models |
| Church et al. (2015) [7] | 1.06 (0.58–1.91) | 0.19 (0.02–1.31) | - | Multivariable analysis | Kaplan-Meier method and compared by the log-rank test |
| Karnezis et al. (2017) [27] | 0.59 (0.21–1.60) | 0.49 (0.12–1.90) | 0.26 (0.04–1.49) | Univariable survival analysis | Kaplan-Meier survival curve |
| Stelloo et al. (2016) [20] | 1.10 (0.39–3.10) | Multivariable analysis | Kaplan-Meier survival analysis | ||
| Bosse et al. (2018) [13] | 0.23 (0.06–0.76) | Multivariable analysis | Kaplan-Meier survival curve | ||
| Pooled HR (95% CI) | 0.90 (0.59–1.38) | 0.41 (0.30–0.55) | 0.23 (0.08–0.64) | ||
| I2 (95% CI, %) | 0.00 (0.00–73.28) | 0.00 (0.00–67.34) | 0.00 (0.00–0.00) |
- 1. Bell DW, Ellenson LH. Molecular genetics of endometrial carcinoma. Annu Rev Pathol 2019; 14: 339-67. ArticlePubMed
- 2. Howlader N, Noone AM, Krapcho M, et al. SEER cancer statistics review, 1975-2011. Bethesda: National Cancer Institute, 2014.
- 3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015; 65: 5-29. ArticlePubMed
- 4. Spurdle AB, Bowman MA, Shamsani J, Kirk J. Endometrial cancer gene panels: clinical diagnostic vs research germline DNA testing. Mod Pathol 2017; 30: 1048-68. ArticlePubMedPDF
- 5. Stelloo E, Nout RA, Naves LC, et al. High concordance of molecular tumor alterations between pre-operative curettage and hysterectomy specimens in patients with endometrial carcinoma. Gynecol Oncol 2014; 133: 197-204. ArticlePubMed
- 6. Hussein YR, Weigelt B, Levine DA, et al. Clinicopathological analysis of endometrial carcinomas harboring somatic POLE exonuclease domain mutations. Mod Pathol 2015; 28: 505-14. ArticlePubMedPDF
- 7. Church DN, Stelloo E, Nout RA, et al. Prognostic significance of POLE proofreading mutations in endometrial cancer. J Natl Cancer Inst 2015; 107: 402.ArticlePubMedPDF
- 8. Cancer Genome Atlas Research Network, Kandoth C, Schultz N, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013; 497: 67-73. ArticlePubMedPMCPDF
- 9. Auguste A, Genestie C, De Bruyn M, et al. Refinement of high-risk endometrial cancer classification using DNA damage response biomarkers: a TransPORTEC initiative. Mod Pathol 2018; 31: 1851-61. ArticlePubMedPDF
- 10. Talhouk A, McConechy MK, Leung S, et al. A clinically applicable molecular-based classification for endometrial cancers. Br J Cancer 2015; 113: 299-310. ArticlePubMedPMCPDF
- 11. Stelloo E, Bosse T, Nout RA, et al. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer: a TransPORTEC initiative. Mod Pathol 2015; 28: 836-44. ArticlePubMedPDF
- 12. Eggink FA, Van Gool IC, Leary A, et al. Immunological profiling of molecularly classified high-risk endometrial cancers identifies POLE-mutant and microsatellite unstable carcinomas as candidates for checkpoint inhibition. Oncoimmunology 2017; 6: e1264565. ArticlePubMedPDF
- 13. Bosse T, Nout RA, McAlpine JN, et al. Molecular classification of grade 3 endometrioid endometrial cancers identifies distinct prognostic subgroups. Am J Surg Pathol 2018; 42: 561-8. ArticlePubMedPMC
- 14. Billingsley CC, Cohn DE, Mutch DG, Stephens JA, Suarez AA, Goodfellow PJ. Polymerase varepsilon (POLE) mutations in endometrial cancer: clinical outcomes and implications for Lynch syndrome testing. Cancer 2015; 121: 386-94. ArticlePubMed
- 15. Le Gallo M, Rudd ML, Urick ME, et al. Somatic mutation profiles of clear cell endometrial tumors revealed by whole exome and targeted gene sequencing. Cancer 2017; 123: 3261-8. ArticlePubMedPMCPDF
- 16. Talhouk A, McConechy MK, Leung S, et al. Confirmation of ProMisE: a simple, genomics-based clinical classifier for endometrial cancer. Cancer 2017; 123: 802-13. ArticlePubMedPDF
- 17. Rosa-Rosa JM, Leskela S, Cristobal-Lana E, et al. Molecular genetic heterogeneity in undifferentiated endometrial carcinomas. Mod Pathol 2016; 29: 1390-8. ArticlePubMedPMCPDF
- 18. Hoang LN, McConechy MK, Meng B, et al. Targeted mutation analysis of endometrial clear cell carcinoma. Histopathology 2015; 66: 664-74. ArticlePubMed
- 19. Abdulfatah E, Wakeling E, Sakr S, et al. Molecular classification of endometrial carcinoma applied to endometrial biopsy specimens: towards early personalized patient management. Gynecol Oncol 2019; 154: 467-74. ArticlePubMed
- 20. Stelloo E, Nout RA, Osse EM, et al. Improved risk assessment by integrating molecular and clinicopathological factors in early-stage endometrial cancer-combined analysis of the PORTEC cohorts. Clin Cancer Res 2016; 22: 4215-24. ArticlePubMedPDF
- 21. Imboden S, Nastic D, Ghaderi M, et al. Phenotype of POLE-mutated endometrial cancer. PLoS One 2019; 14: e0214318. ArticlePubMedPMC
- 22. Billingsley CC, Cohn DE, Mutch DG, Hade EM, Goodfellow PJ. Prognostic significance of POLE exonuclease domain mutations in high-grade endometrioid endometrial cancer on survival and recurrence: a subanalysis. Int J Gynecol Cancer 2016; 26: 933-8. ArticlePubMedPMC
- 23. Talhouk A, Hoang LN, McConechy MK, et al. Molecular classification of endometrial carcinoma on diagnostic specimens is highly concordant with final hysterectomy: earlier prognostic information to guide treatment. Gynecol Oncol 2016; 143: 46-53. ArticlePubMedPMC
- 24. Wong A, Kuick CH, Wong WL, et al. Mutation spectrum of POLE and POLD1 mutations in South East Asian women presenting with grade 3 endometrioid endometrial carcinomas. Gynecol Oncol 2016; 141: 113-20. ArticlePubMed
- 25. Wortman BG, Creutzberg CL, Putter H, et al. Ten-year results of the PORTEC-2 trial for high-intermediate risk endometrial carcinoma: improving patient selection for adjuvant therapy. Br J Cancer 2018; 119: 1067-74. ArticlePubMedPMCPDF
- 26. Kommoss FK, Karnezis AN, Kommoss F, et al. L1CAM further stratifies endometrial carcinoma patients with no specific molecular risk profile. Br J Cancer 2018; 119: 480-6. ArticlePubMedPMCPDF
- 27. Karnezis AN, Leung S, Magrill J, et al. Evaluation of endometrial carcinoma prognostic immunohistochemistry markers in the context of molecular classification. J Pathol Clin Res 2017; 3: 279-93. ArticlePubMedPMCPDF
- 28. Espinosa I, Lee CH, D'Angelo E, Palacios J, Prat J. Undifferentiated and dedifferentiated endometrial carcinomas with POLE exonuclease domain mutations have a favorable prognosis. Am J Surg Pathol 2017; 41: 1121-8. ArticlePubMed
- 29. DeLair DF, Burke KA, Selenica P, et al. The genetic landscape of endometrial clear cell carcinomas. J Pathol 2017; 243: 230-41. ArticlePubMedPMCPDF
- 30. Meng B, Hoang LN, McIntyre JB, et al. POLE exonuclease domain mutation predicts long progression-free survival in grade 3 endometrioid carcinoma of the endometrium. Gynecol Oncol 2014; 134: 15-9. ArticlePubMed
- 31. Bellone S, Bignotti E, Lonardi S, et al. Polymerase epsilon (POLE) ultra-mutation in uterine tumors correlates with T lymphocyte infiltration and increased resistance to platinum-based chemotherapy in vitro. Gynecol Oncol 2017; 144: 146-52. PubMed
- 32. Moher D, Shamseer L, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev 2015; 4: 1.PubMedPMCPDF
- 33. MedCalc statistical software version 13.0.6 [Internet]. Ostend: MedCalc Software, 2014 [cited 2020 Feb 27]. Available from: http://www.medcalc.org.
- 34. Jumaah AS, Al-Haddad HS, Mahdi LH, et al. Increased PTEN gene expression in patients with endometrial carcinoma from areas of high risk depleted uranium exposure. BMC Res Notes 2019; 12: 708.ArticlePubMedPMCPDF
- 35. van Gool IC, Eggink FA, Freeman-Mills L, et al. POLE proofreading mutations elicit an antitumor immune response in endometrial cancer. Clin Cancer Res 2015; 21: 3347-55. ArticlePubMedPMCPDF
- 36. Bakhsh S, Kinloch M, Hoang LN, et al. Histopathological features of endometrial carcinomas associated with POLE mutations: implications for decisions about adjuvant therapy. Histopathology 2016; 68: 916-24. ArticlePubMed
- 37. Sterne JA, Sutton AJ, Ioannidis JP, et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ 2011; 343: d4002.ArticlePubMed
REFERENCES
Figure & Data
References
Citations

- The relationship between histopathological data and molecular alterations with oncological outcomes in endometrioid-type endometrial cancers and a novel POLE mutation
Elif Aksahin, Fuat Demirkiran, Tugan Bese, Sukru Cebi, Abdullah Serdar Acikgoz, Basak Ozge Kayan, Yeliz Aykanat, Ismail Yilmaz, Ayse Namal, Sennur Ilvan, Omer Uysal, Macit Arvas
Journal of Gynecologic Oncology.2026;[Epub] CrossRef - Characterization of Korean Colorectal Cancer Reveals Novel Driver Gene and Clinically Relevant Mutations
Junho Kang, Dong Min Lim, Young‐Joon Kim, Hyeran Shim, Tae‐You Kim, Kyu Joo Park, Sung‐Bum Kang, Chang Sik Yu, Jong Lyul Lee, Yeuni Yu, Hansong Lee, Eun Jung Kwon, Hyo Min Kim, Seongik Mun, Donghee Kwak, Hae Seul Lee, Hye Jin Heo, Eun Kyoung Kim, Seung Eu
MedComm.2026;[Epub] CrossRef - CircRNA-miRNA-mRNA interactome analysis in endometrial cancer
Tikam Chand Dakal, Abhishek Kumar, Pawan Kumar Maurya
Journal of Biomolecular Structure and Dynamics.2025; 43(3): 1486. CrossRef - The prognostic implication of polymerase epsilon-mutated endometrial cancer
Kai-Hung Wang, Dah-Ching Ding
Tzu Chi Medical Journal.2025; 37(2): 135. CrossRef - Functions, interactions and prognostic role of POLE: a bioinformatics analysis
Jonathan Carvajal-Veloza, Fredy Galindo-Morales, Luz Dary Gutierrez-Castañeda
Journal of Gynecologic Oncology.2025;[Epub] CrossRef - Development of Antibody–Drug Conjugates for Malignancies of the Uterine Corpus: A Review
Taro Yamanaka, Tadaaki Nishikawa, Hiroshi Yoshida
Cells.2025; 14(5): 333. CrossRef - Efficacy of dostarlimab in recurrent or advanced mismatch Repair-Deficient endometrial Cancer as a Single-Agent therapy: A systematic review and Meta-Analysis
Ramazan Rezaei, Hedieh Haji Khodaverdi Khani
DARU Journal of Pharmaceutical Sciences.2025;[Epub] CrossRef - POLE-mutated endometrial cancer: new perspectives on the horizon?
Daniele Fanale, Lidia Rita Corsini, Paola Piraino, Erika Pedone, Chiara Brando, Tancredi Didier Bazan Russo, Pietro Ferraro, Alisia Simone, Silvia Contino, Ornella Prestifilippo, Ugo Randazzo, Ambra Giurintano, Carla Ferrante Bannera, Antonio Galvano, Lor
Frontiers in Oncology.2025;[Epub] CrossRef - Mismatch repair, p53, and L1 cell adhesion molecule status influence the response to chemotherapy in advanced and recurrent endometrial cancer
Jung Chul Kim, Byungsoo Ahn, Yong Jae Lee, Eun Ji Nam, Sang Wun Kim, Sunghoon Kim, Young Tae Kim, Eunhyang Park, Jung-Yun Lee
BMC Cancer.2024;[Epub] CrossRef - A single-institution retrospective exploratory analysis on the effectiveness and safety of lenvatinib plus pembrolizumab for advanced endometrial cancer: insights from ProMisE molecular classification system
Yohei Chiba, Masahiro Kagabu, Mitsumasa Osakabe, Rikako Ito, Sho Sato, Eriko Takatori, Yoshitaka Kaido, Takayuki Nagasawa, Tadahiro Shoji, Naoki Yanagawa, Tsukasa Baba
Japanese Journal of Clinical Oncology.2024; 54(4): 424. CrossRef - Potential of molecular classification to guide fertility-sparing management among young patients with endometrial cancer
Nuria Agusti, Alexa Kanbergs, Roni Nitecki
Gynecologic Oncology.2024; 185: 121. CrossRef - Assessing the New 2020 ESGO/ESTRO/ESP Endometrial Cancer Risk Molecular Categorization System for Predicting Survival and Recurrence
Yung-Taek Ouh, Yoonji Oh, Jinwon Joo, Joo Hyun Woo, Hye Jin Han, Hyun Woong Cho, Jae Kwan Lee, Yikyeong Chun, Myoung-nam Lim, Jin Hwa Hong
Cancers.2024; 16(5): 965. CrossRef - The Clinical and Pathological Characteristics of POLE-Mutated Endometrial Cancer: A Comprehensive Review
Xiaohong Yao, Min Feng, Wei Wang
Cancer Management and Research.2024; Volume 16: 117. CrossRef - National Survey of Current Follow-up Protocols for Patients Treated for Endometrial Cancer in the UK
H. Patel, K. Drinkwater, A. Stewart
Clinical Oncology.2024; 36(6): e146. CrossRef - Nab-Paclitaxel-Based Systemic Approach to Achieving Complete Remission for Relapsed Stage III Endometrial Carcinoma: Insights From the Indian Subcontinent
Prasanna Rammohan, Vipulkumar Thummar, Priya Mehta
Cureus.2024;[Epub] CrossRef - High prevalence of “non‐pathogenic” POLE mutation with poor prognosis in a cohort of endometrial cancer from South India
Santhosh Kuriakose, Dhananjayan Dhanasooraj, P. M. Shiny, S. Shammy, V. P. Sona, Anupama A. Manjula, Amrutha Ramachandran, Bindu Vijaykumar, Nayana Susan, M. Dinesan, Uma V. Sankar, Kavitha Ramachandran, P. S. Sreedharan
International Journal of Gynecology & Obstetrics.2024; 166(3): 1263. CrossRef - Patterns and Frequency of Pathogenic Germline Mutations among Patients with Newly-Diagnosed Endometrial Cancer: The Jordanian Exploratory Cancer Genetics (Jo-ECAG) Endometrial Study
Hikmat Abdel-Razeq, Hira Bani Hani, Baha Sharaf, Faris Tamimi, Hanan Khalil, Areej Abu Sheikha, Mais Alkyam, Sarah Abdel-Razeq, Tala Ghatasheh, Tala Radaideh, Suhaib Khater
Cancers.2024; 16(14): 2543. CrossRef - Accelerated clinical response achieved by combining short-term tumor-directed photodynamic therapy with immunotherapy-based systemic therapies in synchronous colorectal cancer with MSI-H and POLE mutation: a case report
Yuhan Wang, Lei Gao, Bin Ma, Jianming Shi, Zhenyu Yin, Weidong Zhu, Hao Chen
Frontiers in Immunology.2024;[Epub] CrossRef - Morphomolecular Correlation and Clinicopathologic Analysis in Endometrial Carcinoma
Göksenil Bülbül, Tekincan Çağri Aktaş, Anil Aysal Ağalar, Safiye Aktaş, Sefa Kurt, Bahadir Saatli, Emine Çağnur Ulukuş
International Journal of Gynecological Pathology.2024; 43(6): 535. CrossRef - Prognostic implications of immunohistochemistry in patients with endometrial cancer
Maria-Bianca Anca-Stanciu, Andrei Manu , Maria Victoria Olinca , Bogdan Cătălin Coroleucă , Diana-Elena Comandaşu , Ciprian Andrei Coroleucă , Călina Maier , Elvira Brătilă
Romanian Journal of Morphology and Embryology.2024; 65(2): 185. CrossRef - Translating biological insights into improved management of endometrial cancer
Jeffrey A. How, Amir A. Jazaeri, Shannon N. Westin, Barrett C. Lawson, Ann H. Klopp, Pamela T. Soliman, Karen H. Lu
Nature Reviews Clinical Oncology.2024; 21(11): 781. CrossRef - Fast and reliable Sanger POLE sequencing protocol in FFPE tissues of endometrial cancer
Izabela Laczmanska, Dagmara Michalowska, Marcin Jedryka, Dorota Blomka, Mariola Semeniuk, Ewelina Czykalko, Mariola Abrahamowska, Paulina Mlynarczykowska, Agnieszka Chrusciel, Ireneusz Pawlak, Adam Maciejczyk
Pathology - Research and Practice.2023; 242: 154315. CrossRef - Uterine Neoplasms, Version 1.2023, NCCN Clinical Practice Guidelines in Oncology
Nadeem Abu-Rustum, Catheryn Yashar, Rebecca Arend, Emma Barber, Kristin Bradley, Rebecca Brooks, Susana M. Campos, Junzo Chino, Hye Sook Chon, Christina Chu, Marta Ann Crispens, Shari Damast, Christine M. Fisher, Peter Frederick, David K. Gaffney, Robert
Journal of the National Comprehensive Cancer Network.2023; 21(2): 181. CrossRef - The hereditary N363K POLE exonuclease mutant extends PPAP tumor spectrum to glioblastomas by causing DNA damage and aneuploidy in addition to increased mismatch mutagenicity
Guillaume Labrousse, Pierre Vande Perre, Genis Parra, Marion Jaffrelot, Laura Leroy, Frederic Chibon, Frederic Escudie, Janick Selves, Jean-Sebastien Hoffmann, Rosine Guimbaud, Malik Lutzmann
NAR Cancer.2023;[Epub] CrossRef - New boundaries for fertility sparing management in endometrial cancer
Alexandros Rodolakis, Vasilis Pergialiotis, Nikolaos Thomakos
Current Opinion in Oncology.2023; 35(5): 394. CrossRef - PD-1 and PD-L1 Blockade plus Chemotherapy in Endometrial Cancer
New England Journal of Medicine.2023; 389(9): 866. CrossRef - The Shifting Landscape of p53abn Endometrial Cancers: A Review of the Prognostic and Predictive Impact and Current Therapeutic Directions
Angelo Anater
Journal of Medical and Radiation Oncology.2023; 3(2): 1. CrossRef - The Advantages of Next-Generation Sequencing Molecular Classification in Endometrial Cancer Diagnosis
Daniela Rivera, Michele Paudice, Giulia Accorsi, Floriana Valentino, Marta Ingaliso, Ada Pianezzi, Paola Roggieri, Lucia Trevisan, Giulia Buzzatti, Serafina Mammoliti, Fabio Barra, Simone Ferrero, Gabriella Cirmena, Viviana Gismondi, Valerio Gaetano Vello
Journal of Clinical Medicine.2023; 12(23): 7236. CrossRef - The clinicopathology and survival characteristics of patients with POLE proofreading mutations in endometrial carcinoma: A systematic review and meta-analysis
Alaa Salah Jumaah, Hawraa Sahib Al-Haddad, Katherine Ann McAllister, Akeel Abed Yasseen, Manish S. Patankar
PLOS ONE.2022; 17(2): e0263585. CrossRef - Enhanced polymerase activity permits efficient synthesis by cancer-associated DNA polymerase ϵ variants at low dNTP levels
Stephanie R Barbari, Annette K Beach, Joel G Markgren, Vimal Parkash, Elizabeth A Moore, Erik Johansson, Polina V Shcherbakova
Nucleic Acids Research.2022; 50(14): 8023. CrossRef - The Role of Immunohistochemistry Markers in Endometrial Cancer with Mismatch Repair Deficiency: A Systematic Review
Amelia Favier, Justine Varinot, Catherine Uzan, Alex Duval, Isabelle Brocheriou, Geoffroy Canlorbe
Cancers.2022; 14(15): 3783. CrossRef - The clinicopathological characteristics of POLE-mutated/ultramutated endometrial carcinoma and prognostic value of POLE status: a meta-analysis based on 49 articles incorporating 12,120 patients
Qing Wu, Nianhai Zhang, Xianhe Xie
BMC Cancer.2022;[Epub] CrossRef - Mismatch repair deficiency and clinicopathological characteristics in endometrial carcinoma: a systematic review and meta-analysis
Alaa Salah Jumaah, Hawraa Sahib Al-Haddad, Mais Muhammed Salem, Katherine Ann McAllister, Akeel Abed Yasseen
Journal of Pathology and Translational Medicine.2021; 55(3): 202. CrossRef - Evaluation of treatment effects in patients with endometrial cancer and POLE mutations: An individual patient data meta‐analysis
Jessica N. McAlpine, Derek S. Chiu, Remi A. Nout, David N. Church, Pascal Schmidt, Stephanie Lam, Samuel Leung, Stefania Bellone, Adele Wong, Sara Y. Brucker, Cheng Han Lee, Blaise A. Clarke, David G. Huntsman, Marcus Q. Bernardini, Joanne Ngeow, Alessand
Cancer.2021; 127(14): 2409. CrossRef - Endometrial cancer
Vicky Makker, Helen MacKay, Isabelle Ray-Coquard, Douglas A. Levine, Shannon N. Westin, Daisuke Aoki, Ana Oaknin
Nature Reviews Disease Primers.2021;[Epub] CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure



Fig. 1.
Fig. 2.
Fig. 3.
| Study | EC cohort size | Proportion POLE-mutant | Country | Sequencing method | Location of exonuclease mutations |
|---|---|---|---|---|---|
| The Cancer Genome Atlas Research Network (2013) [8] | 248 | 17 | USA | Exome sequencing | Hotspots: Pro286Arg and Val411Leu |
| Auguste et al. (2018) [9] | 102 | 9 | Canada and Europe | Sanger sequencing | Exons 9 and 13 |
| Talhouk et al. (2015) [10] | 143 | 12 | Canada | Fluidigm-MiSeq and sanger sequencing | Exons 9–14 |
| Stelloo et al. (2015) [11] | 116 | 14 | Europe and Australia | Sanger sequencing | Exons 9 and 13 |
| Eggink et al. (2017) [12] | 116 | 15 | Europe and Australia | Sanger sequencing | Exons 9, 13 and 14 |
| Wortman et al. (2018) [25] | 344 | 16 | Netherlands | Sequencing | Not reported |
| Kommoss et al. (2018) [26] | 452 | 42 | Germany | Sequencing | Exons 9–14 |
| Bosse et al. (2018) [13] | 376 | 48 | USA, Canada, and Europe | Sanger or next-generation approaches | Hotspots in the exonuclease domain (exons 9–14) |
| Billingsley et al. (2015) [14] | 535 | 30 | USA | Sanger sequencing | Residues 268–471 |
| Le Gallo et al. (2017) [15] | 63 | 0 | USA and Europe | Sanger sequencing | Not reported |
| Karnezis et al. (2017) [27] | 460 | 42 | Canada | Sequencing | Not reported |
| Talhouk et al. (2017) [16] | 319 | 30 | Canada | Sanger sequencing | Exons 9–14 |
| Rosa-Rosa et al. (2016) [17] | 18 | 2 | USA and Europe | Sanger sequencing | Exons 9 and 13 |
| Wortman et al. (2018) [25] | 416 | 16 | Netherlands | Sequencing | Not reported |
| Espinosa et al. (2017) [28] | 21 | 9 | Spain | Sequencing | Exons 9 to 14 |
| Stelloo et al. (2015) [11] | 116 | 14 | Europe | Sanger sequencing | Exons 9 and 13 |
| Hoang et al. (2015) [18] | 14 | 0 | Canada | Sanger sequencing | Exons 9–14 |
| DeLair et al. (2017) [29] | 30 | 2 | USA | Sequencing | Exons 9–14 |
| Abdulfatah et al. (2019) [19] | 60 | 2 | USA | Sanger sequencing | Exons 9 and 13 |
| Wong et al. (2016) [24] | 47 | 14 | Singapore | Next generation sequencing | Exons 9–14 |
| Stelloo et al. (2016) [20] | 834 | 49 | Netherlands | Sanger sequencing | Exons 9 and 13 |
| Imboden et al. (2019) [21] | 599 | 38 | Sweden | Sanger sequencing | Exons 9–14 |
| Church et al. (2015) [7] | 788 | 48 | Europe | Sanger sequencing | Exons 9 and 13 |
| Billingsley et al. (2016) [22] | 72 | 7 | USA | Sanger sequencing | Residues 268–471 |
| Talhouk et al. (2016) [23] | 57 | 10 | USA and Canada | Ultra-deep MiSeq or sanger sequencing | Exons 9–14 |
| Clinicopathological characteristics in EC | Pooled % portion (95% CI, %) | No. of studies | I2 (95% CI) | p-value | Model |
|---|---|---|---|---|---|
| Overall POLE mutation | 8.59 (7.01–10.32) | 25 | 78.10 (68.15–84.94) | < .001 | Random effect |
| POLE mutation in type I | 8.22 (6.27–10.42) | 9 | 74.88 (51.43–87.00) | < .001 | Random effect |
| POLE mutation in type II | 0.93 (0.34–1.81) | 10 | 75.32 (54.08–86.74) | < .001 | Random effect |
| Stage I–II | 89.51 (81.11–95.66) | 10 | 69.09 (40.43–83.96) | < .001 | Random effect |
| Stage III–IV | 14.77 (5.99–26.59) | 7 | 65.96 (23.79–84.79) | < .001 | Random effect |
| Grade I–II | 46.36 (30.66–62.43) | 7 | 82.15 (64.34–91.06) | < .001 | Random effect |
| Grade III | 51.53 (36.08–66.84) | 8 | 81.79 (65.23–90.46) | < .001 | Random effect |
| Lymphovascular invasion | 31.11 (10.44–56.86) | 8 | 93.34 (89.15–95.91) | < .001 | Random effect |
| Myometrial invasion less than 50% | 49.90 (43.71–56.21) | 7 | 22.10 (0.00–65.16) | 0.260 | Fixed effect |
| Clinicopathology: POLE-mutant vs. wild type | Pooled odd ratio (95% CI) | No. of studies | I2 (95% CI, %) | p-value for I2 | Model |
|---|---|---|---|---|---|
| Stage I–II EC | 3.727 (2.063–6.732) | 8 | 0.00 (0.00–25.07) | .890 | Fixed effect |
| Stage III–IV EC | 0.269 (0.147–0.494) | 7 | 0.00 (0.00–53.51) | .716 | Fixed effect |
| Grade I–II EC | 0.400 (0.295–0.542) | 8 | 3.95 (0.00–69.18) | .399 | Fixed effect |
| Grade III EC | 2.246 (1.655–3.048) | 8 | 0.00 (0.00–29.91) | .865 | Fixed effect |
| LVI | 0.929 (0.643–1.341) | 8 | 6.95 (0.00–70.15) | .376 | Fixed effect |
| MI less than 50% | 1.481 (0.996–2.202) | 6 | 47.63 (0.00–79.24) | .089 | Random effect |
| Type I endometrioid histology | 1.721 (1.113–2.662) | 9 | 0.00 (0.00–68.45) | .486 | Fixed effect |
| Study | OS estimated HR (95% CI) | DSS estimated HR (95% CI) | PFS estimated HR (95% CI) | Survival analysis test | Method |
|---|---|---|---|---|---|
| Talhouk et al. (2017) [16] | 1.01 (0.29–3.42) | 0.42 (0.30–0.57) | - | Multivariable analysis | Kaplan-Meier survival analysis |
| Talhouk et al. (2015) [10] | 0.17 (0.01–1.98) | 0.170 (0.01–1.99) | - | Multivariable analysis | Kaplan-Meier with log-rank significance testing and Cox proportional hazard regression models |
| Church et al. (2015) [7] | 1.06 (0.58–1.91) | 0.19 (0.02–1.31) | - | Multivariable analysis | Kaplan-Meier method and compared by the log-rank test |
| Karnezis et al. (2017) [27] | 0.59 (0.21–1.60) | 0.49 (0.12–1.90) | 0.26 (0.04–1.49) | Univariable survival analysis | Kaplan-Meier survival curve |
| Stelloo et al. (2016) [20] | 1.10 (0.39–3.10) | Multivariable analysis | Kaplan-Meier survival analysis | ||
| Bosse et al. (2018) [13] | 0.23 (0.06–0.76) | Multivariable analysis | Kaplan-Meier survival curve | ||
| Pooled HR (95% CI) | 0.90 (0.59–1.38) | 0.41 (0.30–0.55) | 0.23 (0.08–0.64) | ||
| I2 (95% CI, %) | 0.00 (0.00–73.28) | 0.00 (0.00–67.34) | 0.00 (0.00–0.00) |

 E-submission
E-submission