Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 55(3); 2021 > Article
-
Original Article
Mismatch repair deficiency and clinicopathological characteristics in endometrial carcinoma: a systematic review and meta-analysis -
Alaa Salah Jumaah1
 , Hawraa Sahib Al-Haddad2
, Hawraa Sahib Al-Haddad2 , Mais Muhammed Salem1
, Mais Muhammed Salem1 , Katherine Ann McAllister3
, Katherine Ann McAllister3 , Akeel Abed Yasseen1
, Akeel Abed Yasseen1
-
Journal of Pathology and Translational Medicine 2021;55(3):202-211.
DOI: https://doi.org/10.4132/jptm.2021.02.19
Published online: April 14, 2021
1Department of Pathology and Forensic Medicine, Faculty of Medicine, University of Kufa, Kufa, Iraq
2Al-Furat Al-Awsat Hospital, Kufa, Iraq
3School of Biomedical Sciences, Ulster University, Northern Ireland, UK
- Corresponding Author: Akeel Abed Yasseen, PhD, Department of Pathology and Forensic Medicine, Faculty of Medicine, University of Kufa, Kufa, P.O. Box 21, Najaf Governorate, Iraq Tel: +9647811131586, E-mail: akeelyasseen@uokufa.edu.iq
© 2021 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Figure & Data
References
Citations

- Molecular Classification and Clinical Outcomes in Endometrial Cancer: Real-World Evidence from a Tertiary Care Center
Tanadon Salakphet, Prapaporn Suprasert, Tip Pongsuvareeyakul, Chinachote Teerapakpinyo, Surapan Khunamornpong
Cancers.2026; 18(2): 181. CrossRef - Comparison of Molecular Alterations and Survival Analysis in Uterine Endometrioid Carcinomas and Serous Carcinomas: An In Silico Study
Zeynep Sağnak Yılmaz
Archives of Current Medical Research.2026; 7(1): 244. CrossRef - Prevalence of Mismatch Repair Gene Defects by Means of Immuno-histochemistry Staining for MMR Proteins in Endometrial Cancer
Kaustubh Girish Burde, Indu R. Nair, Pavithran Keechilattu, Anupama Rajanbabu
The Journal of Obstetrics and Gynecology of India.2025; 75(S1): 135. CrossRef - Deficient Mismatch Repair and Microsatellite Instability in Solid Tumors
Joy A. Awosika, James L. Gulley, Danielle M. Pastor
International Journal of Molecular Sciences.2025; 26(9): 4394. CrossRef - Changes in Nucleolar Activity Under Conditions of Microsatellite Instability in the Uterine Mucosa in Precancer and Endometrial Cancer
A. V. Zatvornickaya, E. L. Kazachkov, E. A. Kazachkova
Ural Medical Journal.2025; 24(2): 71. CrossRef - Current approaches to first-line therapy for endometrial cancer: the role of lenvatinib and pembrolizumab combination therapy
V. M. Nechushkina, S. V. Khokhlova, D. A. Nosov, E. A. Ulrikh, L. A. Kolomiets, A. A. Rumyantsev, G. A. Raskin
Tumors of female reproductive system.2025; 21(2): 127. CrossRef - Endometrial carcinoma in Kenya: clinical and biomarker profiles of 123 cases seen at two tertiary referral centers
Olivia Chesikaw, Allan Njau, Erick Chesori, Khadija Warfa, Anisa Mburu, Afrin Fatima Shaffi, Natalie Banet, Jonathan Wawire
Frontiers in Medicine.2025;[Epub] CrossRef - Association Between MMR Status and Prognostic Pathological Factors in Endometrioid Endometrial Cancer—A Single-Center Retrospective Study
Cezary Miedziarek, Hubert Bochyński, Katarzyna Bociańska, Michał Potograbski, Piotr Tyburski, Mikołaj Piotr Zaborowski, Ewa Nowak-Markwitz
Cancers.2025; 17(22): 3605. CrossRef - Mismatch Repair Deficiency Profiling and Its Impact on Management and Prognosis in Endometrial Cancer Patients: A Comprehensive Update
Emmanouela-Aliki Almperi, Chrysoula Margioula-Siarkou, Aristarchos Almperis, Georgia Margioula-Siarkou, Stefanos Flindris, Alexandros I Daponte, Theodora Papamitsou, Konstantinos Dinas, Stamatios Petousis
Cureus.2025;[Epub] CrossRef - Mismatch Repair Deficiency as a Predictor of Lymph Node Metastasis in Endometrioid Endometrial Carcinoma
Esra Keles, Mustafa Maraşlı, Meziha Taşyürek Coşkun, Elif Ünlügedik Sayın, Öncel İpekçi, Mervernur Şahin, Fatih Şanlıkan, Uğur Kemal Öztürk, İsmail Bağlar, Sahra Sultan Kara, Melis Hazırlar
Muğla Sıtkı Koçman Üniversitesi Tıp Dergisi.2025; 12(3): 269. CrossRef - Guidelines of the Brazilian Society of Surgical Oncology for anatomopathological, immunohistochemical, and molecular testing in female tumors
Reitan Ribeiro, Filomena Marino Carvalho, Glauco Baiocchi, Rodrigo Santa Cruz Guindalini, Juliano Rodrigues da Cunha, Carlos Henrique dos Anjos, Caroline de Nadai Costa, Ana Carolina Leite Vieira Costa Gifoni, Renato Cagnacci Neto, Allyne Queiroz Carneiro
Journal of Surgical Oncology.2024; 130(4): 882. CrossRef - Microsatellite instability as a reliable marker of coexisting endometrial cancer in atypical endometrial hyperplasia
А. E. Protasova, G. A. Raskin, M. S. Sobivchak
Tumors of female reproductive system.2024; 20(2): 105. CrossRef - Refining of cancer-specific genes in microsatellite-unstable colon and endometrial cancers using modified partial least square discriminant analysis
Woong Na, Sung Hak Lee, Seunghee Lee, Jong-Seok Kim, Seung Yun Han, Yong Min Kim, Mihye Kwon, Young Soo Song
Medicine.2024; 103(52): e41134. CrossRef - Cancer-specific functional profiling in microsatellite-unstable (MSI) colon and endometrial cancers using combined differentially expressed genes and biclustering analysis
Woong Na, Il Ju Lee, Insong Koh, Mihye Kwon, Young Soo Song, Sung Hak Lee
Medicine.2023; 102(19): e33647. CrossRef - Clinicopathological characteristics of endometrial carcinomas according to DNA mismatch repair protein status
Daniela de Freitas, Fernando Nalesso Aguiar, Cristina Anton, Danielle Cristina de Almeida, Carlos Eduardo Bacchi, Jesus Paula Carvalho, Filomena Marino Carvalho
Heliyon.2023; 9(6): e17495. CrossRef - Mesonephric-like Adenocarcinoma of the Uterine Corpus: Genomic and Immunohistochemical Profiling with Comprehensive Clinicopathological Analysis of 17 Consecutive Cases from a Single Institution
Hyun-Hee Koh, Eunhyang Park, Hyun-Soo Kim
Biomedicines.2023; 11(8): 2269. CrossRef - miR-486-3p Controls the Apoptosis of Endometrial Carcinoma Cells
Donghua Wang, Xiaoli Liu, Lirong Cao, Shixiong Gong, Yi He, Xiangbin Jiang, Zhongxian Wang
Journal of Biomaterials and Tissue Engineering.2022; 12(5): 1002. CrossRef - The Role of Immunohistochemistry Markers in Endometrial Cancer with Mismatch Repair Deficiency: A Systematic Review
Amelia Favier, Justine Varinot, Catherine Uzan, Alex Duval, Isabelle Brocheriou, Geoffroy Canlorbe
Cancers.2022; 14(15): 3783. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure
- Related articles
-
- AMACR is a highly sensitive and specific immunohistochemical marker for diagnosing prostate cancer on biopsy: a systematic review and meta-analysis
- Thoracic aortic calcification as a predictor of coronary artery disease: a systematic review and meta-analysis
- Prognostic and clinicopathological significance of Fusobacterium nucleatum in colorectal cancer: a systemic review and meta-analysis



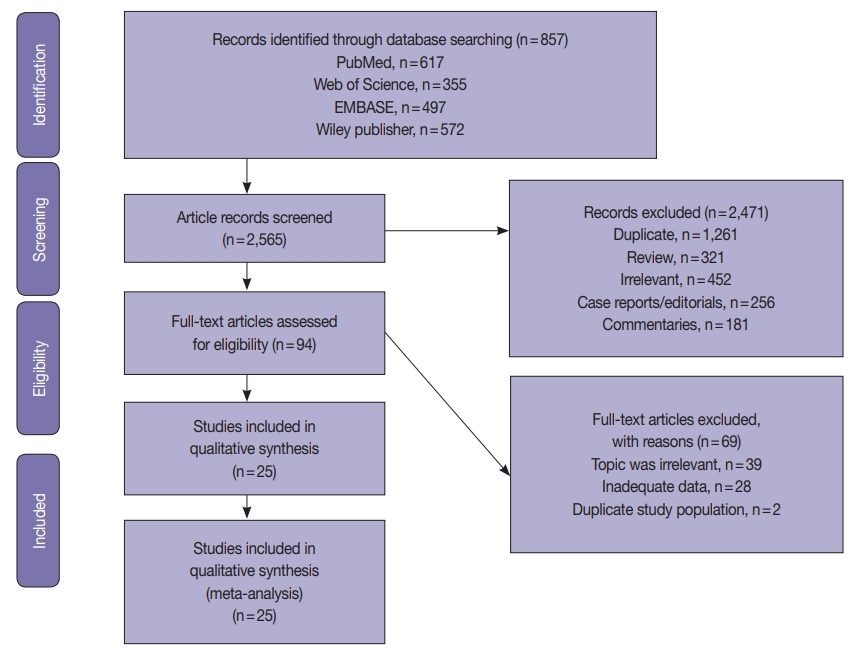
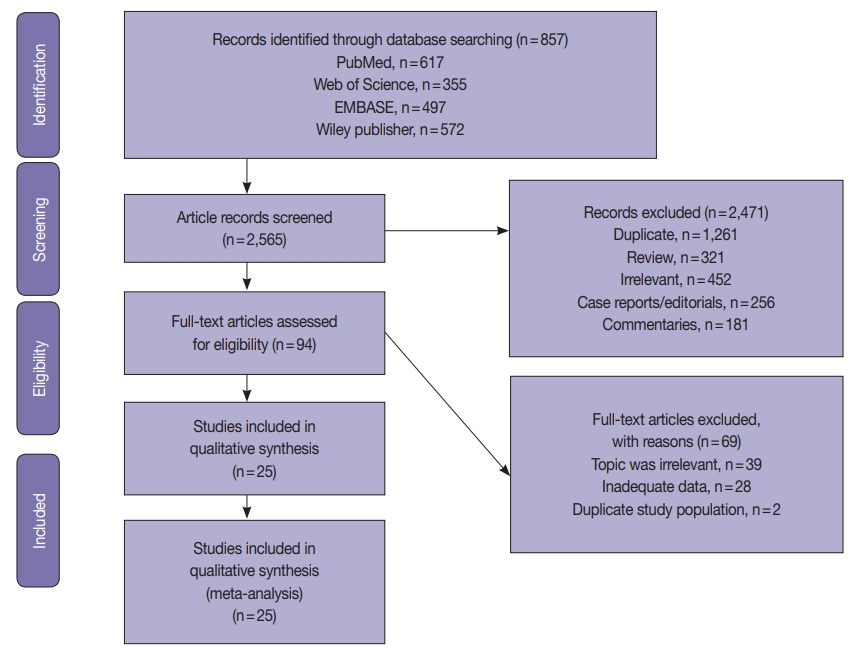
Fig. 1.
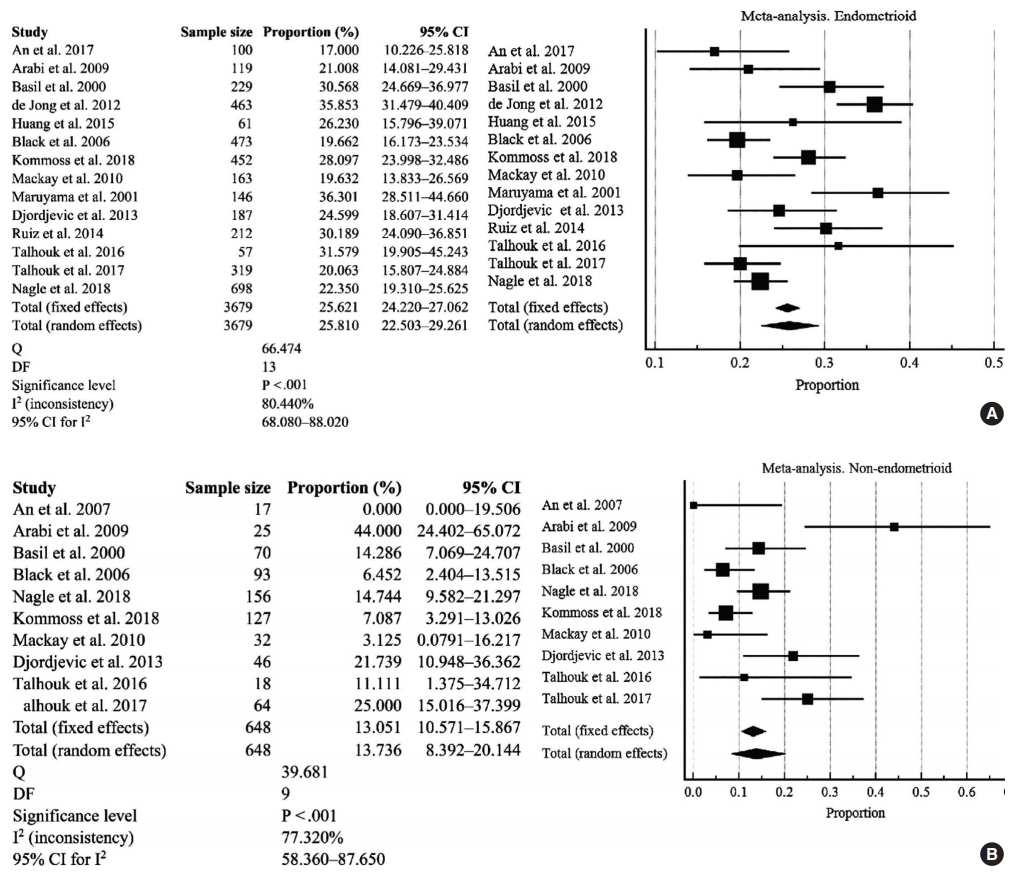
Fig. 2.
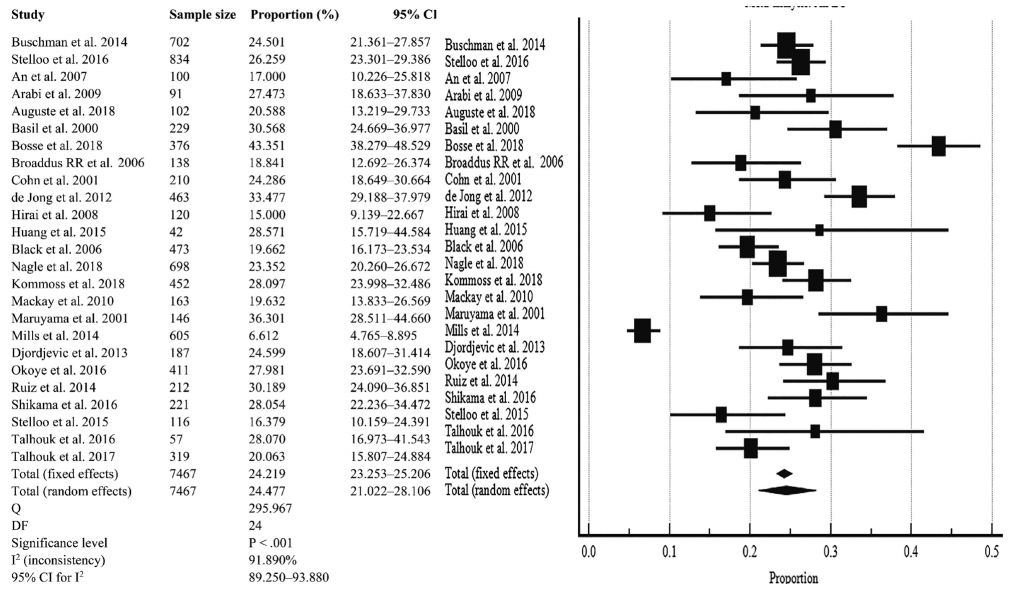
Fig. 3.
| Study | Sample size | D-MMR EC (%) | Study site | Detection method and D-MMR marker | Clinicopathologic characteristics |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| D-MMR subjects Grade I-II EC | D-MMR Grade III EC | D-MMR Stage I-II EC | D-MMR Stage III-VI EC | D-MMR subjects with MI < 50% |
D-MMR subjects with MI > 50% |
D-MMR EC with positive LVI | |||||
| Buchanan et al. (2014) [6] | 702 | 24.501 | Australia | IHC amarkers: MLH1, MSH2, MSH6, PMS2 (protein loss or presence) | NR | NR | NR | NR | NR | NR | NR |
| Stelloo et al. (2016) [7] | 834 | 26.259 | Nether-lands | IHC markers: MLH1, MSH2, MSH6, PMS2 proteins | NR | NR | NR | NR | 148 | 71 | 19 |
| An et al. (2007) [8] | 93 | 18.280 | South Korea | PCR | 12 | 5 | NR | NR | 7 | 10 | 7 |
| Arabi et al. (2009) [9] | 91 | 27.473 | USA | IHC markers: MLH1, MSH2, MSH6 proteins | NR | NR | 13 | 12 | 15 | 10 | 14 |
| Auguste et al. (2018) [10] | 102 | 20.588 | USA, Canada | IHC markers: MLH1, MLH2, MSH6, PMS2 | NR | NR | NR | NR | NR | NR | NR |
| Basil et al. (2000) [11] | 229 | 30.568 | USA | PCR markers: BAT26, BAT25, D5S346, D2S123, D17S250 | NR | NR | 61 | 9 | NR | NR | NR |
| Bosse et al. (2018) [12] | 376 | 43.351 | USA, Canada, UK | IHC markers: MLH1, MSH2, MSH6, PMSU proteins | NR | NR | NR | NR | NR | NR | NR |
| Broaddus et al. (2006) [13] | 138 | 18.841 | USA | IHC markers: MLH1 and MSH2 (n = 50) sequencing of MLH1 and MSH2 genes | NR | NR | 16 | 10 | NR | NR | 14 |
| Cohn et al. (2001) [14] | 210 | 24.286 | USA | PCR markers: BAT-25, BAT-26, D5S346, D2S123, D17S250 | NR | NR | NR | NR | NR | NR | NR |
| de Jong et al. (2012) [15] | 463 | 33.477 | Netherlands | IHC markers: MLH1, MSH2, MSH6 proteins | NR | NR | NR | NR | NR | NR | 98 |
| Hirai et al. (2008) [16] | 120 | 15.000 | Japan | Sequencing of MLH1, MLH2, MSH6 genes | NR | NR | NR | NR | NR | NR | NR |
| Huang et al. (2015) [17] | 42 | 28.571 | Taiwan | IHC (MLH1, PMS2, MSH6, and MSH2), PCR (BAT-25, BAT-26, NR-21, NR-24, and MONO-27) | NR | NR | NR | NR | NR | NR | NR |
| Black et al. (2006) [18] | 473 | 19.662 | USA | PCR markers: BAT25, BAT26, D2S123M, D5S346, D17S250 | 72 | 21 | 77 | 16 | 67 | 25 | 30 |
| Nagle et al. (2018) [19] | 698 | 19.198 | Australia | IHC (MLH1, MSH2, MSH6, PMS2), MLH1 methylation testing was conducted for all cases with MLH1/PMS2 loss | NR | NR | 125 | 17 | NR | NR | 47 |
| Kommoss et al. (2018) [20] | 452 | 28.097 | USA, Canada, Germany | IHC to detect MMR protein loss in markers PMS2 and MSH6 | 102 | 25 | NR | NR | 44 | 58 | 25 |
| Mackay et al. (2010) [21] | 163 | 19.632 | Canada | PCR testing of MSI, markers are BAT25/26 | 21 | 11 | NR | NR | NR | NR | NR |
| Maruyama et al. (2001) [22] | 146 | 36.301 | Japan | IHC marker MSH2, MLH1 | NR | NR | NR | NR | NR | NR | NR |
| Mills et al. (2014) [23] | 605 | 6.612 | USA | IHC of MLH1, MSHU, MSH6, PMS2 proteins, PCR confirmation of MLH1/PMS2 DNA methylation | 26 | 9 | NR | NR | NR | NR | NR |
| Djordjevic et al. (2013) [24] | 186 | 24.731 | USA | IHC detection of MLH1, MSH2, MSH6, PMS2 proteins, PCR confirmation of MLH1 methylation to confirm loss of MLH1 protein | 28 | 8 | NR | NR | NR | NR | NR |
| Okoye et al. (2016) [25] | 411 | 27.981 | USA | IHC to detect loss of MLH1, MSH2, MSH6, PMS2, PCR confirmation of MLH1 methylation | NR | NR | NR | NR | NR | NR | NR |
| Ruiz et al. (2014) [26] | 212 | 30.189 | Spain | IHC of MLH1, MSH2, MSH6, PMS2 markers | 58 | 16 | 50 | 10 | 50 | 14 | 6 |
| Shikama et al. (2016) [27] | 221 | 28.054 | Japan | IHC of MMR proteins MLH1, MSH2, MSH6, PMS2 | NR | NR | 39 | 18 | NR | NR | 27 |
| PCR confirmation of MLH1 methylation | |||||||||||
| Stelloo et al. (2015) [28] | 116 | 16.379 | Europe | IHC of MLH1, MSH2, MSH6, PMS2, PCR MLH1 methylation confirmation | NR | NR | NR | NR | NR | NR | NR |
| Promega (ver. 1.2) PCR testing of MSI markers | |||||||||||
| Talhouk et al. (2016) [29] | 57 | 31.579 | Canada, USA | IHC of MMR proteins MLH1, MSH2, MSH6, PMS2 | 10 | 6 | 9 | 3 | 6 | 5 | 7 |
| Talhouk et al. (2017) [30] | 319 | 9.718 | Canada | IHC for presence or absence MMR proteins MLH1, MSH2, MSH6, PMS2 | NR | NR | NR | NR | NR | NR | 28 |
| Clinicopathological characteristics in EC | Pooled % portion (95% CI) | No. of studies | I2 (95% CI, %) | p-value | Model |
|---|---|---|---|---|---|
| Overall D-MMR mutation | 24.477 (21.022–28.106) | 25 | 91.890 (89.250–93.880) | < .001 | Random effect |
| D-MMR mutation in type I | 25.810 (22.503–29.261) | 14 | 80.440 (68.080–88.020) | < .001 | Random effect |
| D-MMR mutation in type II | 13.736 (8.392–20.144) | 10 | 77.320 (58.360–87.650) | < .001 | Random effect |
| Stage I–II | 79.430 (71.500–86.357) | 6 | 68.640 (25.980–86.710) | .007 | Random effect |
| Stage III–IV | 20.168 (13.746–27.469) | 6 | 61.850 (7.020–84.350) | .022 | Random effect |
| Grade I–II | 65.718 (52.602–77.714) | 10 | 91.700 (86.860–94.760) | < .001 | Random effect |
| Grade III | 21.529 (15.930–27.718) | 10 | 94.050 (90.97–96.08) | < .001 | Random effect |
| Lymphovascular invasion | 32.105 (21.371–43.896) | 10 | 91.380 (86.270–94.590) | < .001 | Random effect |
| MI less than 50% | 51.807 (38.514–64.971) | 8 | 89.860 (82.410–94.150) | < .001 | Random effect |
| MI more than 50% | 42.346 (28.576–56.750) | 8 | 91.360 (85.380–94.890) | < .001 | Random effect |
| Clinicopathology D-MMR EC vs. wild type | Pooled odds ratio (95% CI) | No. of studies | I2 (95% CI, %) | p-value for I2 | Model |
|---|---|---|---|---|---|
| Stage I–II EC | 1.565 (0.894–2.740) | 6 | 70.100 (30.040–87.220) | .005 | Random effect |
| Stage III–V EC | 0.936 (0.593–1.478) | 6 | 51.210 (0.000–80.580) | .068 | Random effect |
| Grade I–II EC | 0.706 (0.257–1.940) | 9 | 94.400 (91.360–96.370) | < .001 | Random effect |
| Grade III EC | 1.384 (0.806–2.375) | 7 | 69.430 (32.800–86.100) | .003 | Random effect |
| LVI | 1.765 (1.293–2.409) | 10 | 51.590 (0.530–76.440) | .028 | Random effect |
| MI less than 50% | 1.230 (0.849–1.782) | 8 | 65.900 (27.620–83.930) | .004 | Random effect |
| MI more than 50% | 1.271 (0.871–1.853) | 8 | 65.750 (27.250–83.870) | .004 | Random effect |
| Type I endometrioid histology | 1.389 (0.519–3.720) | 10 | 92.130 (87.630–95.000 | < .001 | Random effect |
| Type II non-endometrioid histology | 0.450 (0.349–0.579) | 10 | 31.500 (0.000–67.300) | .156 | Fixed effect |
D-MMR, MMR deficiency; EC, endometrial carcinoma; MI, myometrial invasion; LVI, lymphovascular invasion; IHC, immunohistochemistry; NR, not reported; PCR, polymerase chain reaction; MMR, mismatch repair; MSI, microsatellite instability. MI < 50%: MI is less than 50% of the total myometrial thickness; MI > 50%: MI is greater than 50% of the total myometrial thickness.
D-MMR, mismatch repair deficiency; EC, endometrial carcinoma; CI, confidence interval; MI, myometrial invasion.
D-MMR, mismatch repair deficiency; EC, endometrial carcinoma; CI, confidence interval; LVI, lymphovascular invasion; MI, myometrial invasion.

 E-submission
E-submission