Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 56(6); 2022 > Article
-
Original Article
Usefulness of BRAF VE1 immunohistochemistry in non–small cell lung cancers: a multi-institutional study by 15 pathologists in Korea -
Sunhee Chang1
 , Yoon-La Choi2
, Yoon-La Choi2 , Hyo Sup Shim3
, Hyo Sup Shim3 , Geon Kook Lee4
, Geon Kook Lee4 , Seung Yeon Ha5
, Seung Yeon Ha5 , Korean Cardiopulmonary Pathology Study Group
, Korean Cardiopulmonary Pathology Study Group -
Journal of Pathology and Translational Medicine 2022;56(6):334-341.
DOI: https://doi.org/10.4132/jptm.2022.08.22
Published online: October 27, 2022
1Department of Pathology, Inje University Ilsan Paik Hospital, Goyang, Korea
2Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
3Department of Pathology, Yonsei University College of Medicine, Seoul, Korea
4Department of Pathology, National Cancer Center, Goyang, Korea
5Department of Pathology, Gachon University Gil Medical Center, Gachon University College of Medicine, Incheon, Korea
-
Corresponding Author: Geon Kook Lee, MD, Department of Pathology, National Cancer Center, 323 Ilsan-ro, Ilsandong-gu, Goyang 10408, Korea Tel: +82-31-920-1746, Fax: +82-31-920-1369, E-mail: gklee@ncc.re.kr
Corresponding Author: Seung Yeon Ha, MD, Department of Pathology, Gachon University Gil Medical Center, Gachon University College of Medicine, 21 Namdong-daero 774beon-gil, Namdong-gu, Incheon 21565, Korea Tel: +82-32-460-3078, Fax: +82-32-460-2394, E-mail: syha@gilhospital.com
© 2022 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- Next-generation sequencing (NGS) is an approved test to select patients for BRAF V600E targeted therapy in Korea. However, the high cost, long turnaround times, and the need for sophisticated equipment and skilled personnel limit the use of NGS in daily practice. Immunohistochemistry (IHC) is a rapid and relatively inexpensive assay available in most laboratories. Therefore, in this study, we evaluate the usefulness of BRAF VE1 IHC in terms of predictive value and interobserver agreement in non–small cell lung cancers (NSCLCs).
-
Methods
- A total of 30 cases with known BRAF mutation status were selected, including 20 cases of lung adenocarcinomas, six cases of colorectal adenocarcinomas, and four cases of papillary thyroid carcinomas. IHC for BRAF V600E was carried out using the VE1 antibody. Fifteen pathologists independently scored both the staining intensity and the percentage of tumor cell staining on whole slide images.
-
Results
- In the lung adenocarcinoma subset, interobserver agreement for the percentage of tumor cell staining and staining intensity was good (percentage of tumor cell staining, intraclass correlation coefficient = 0.869; staining intensity, kappa = 0.849). The interobserver agreement for the interpretation using the cutoff of 40% was almost perfect in the entire study group and the lung adenocarcinoma subset (kappa = 0.815). Sensitivity, specificity, positive predictive value, and negative predictive value of BRAF VE1 IHC were 80.0%, 90.0%, 88.9%, and 81.8%, respectively.
-
Conclusions
- BRAF VE1 IHC could be a screening test for the detection of BRAF V600E mutation in NSCLC. However, further studies are needed to optimize the protocol and to establish and validate interpretation criteria for BRAF VE1 IHC.
- Materials
- A total of 30 cases with known BRAF mutation status were selected from the archives of the Department of Pathology of Samsung Medical Center. Of these, 20 cases were lung adenocarcinomas (15 resections, 3 endobronchial ultrasound [EBUS]–guided biopsies, one needle biopsy, and one bronchoscopic biopsy), and six were colon adenocarcinomas, and four were PTCs (Table 2). The BRAF V600E mutation status was examined by real-time polymerase chain reaction using the Real-Q BRAF V600E detection kit (Biosewoom, Seoul, Korea) and a BRAF probe and primer mixture according to the manufacturer’s protocol. Twenty of 30 cases had BRAF V600E mutation. Of 20 cases of lung adenocarcinoma, 10 cases were positive for BRAF V600E mutation, and others were negative. All six colon adenocarcinomas and four PTCs were positive for BRAF V600E mutation.
- Immunohistochemical staining and scoring
- IHC for BRAF V600E was carried out using Ventana BenchMark ULTRA IHC/ISH (Ventana Medical Systems, Tucson, AZ, USA) immunostainer. Unstained slides were prepared by cutting 4-μm-thick sections. Antigen retrieval was performed using ULTRA Cell Conditioning Solution (Ventana Medical Systems). The sections were incubated with the VE1 primary antibody (mouse monoclonal, prediluted, Ventana Medical Systems) for 16 minutes at 36°C. The slides were visualized using OptiView DAB IHC Detection Kit (Ventana Medical Systems), followed by hematoxylin II counterstaining. PTCs with BRAF V600E mutation were used as positive controls. To obtain whole slide images, the IHC slides were scanned with DP-200 (Roche Diagnostics, Risch-Rotkreuz, Switzerland).
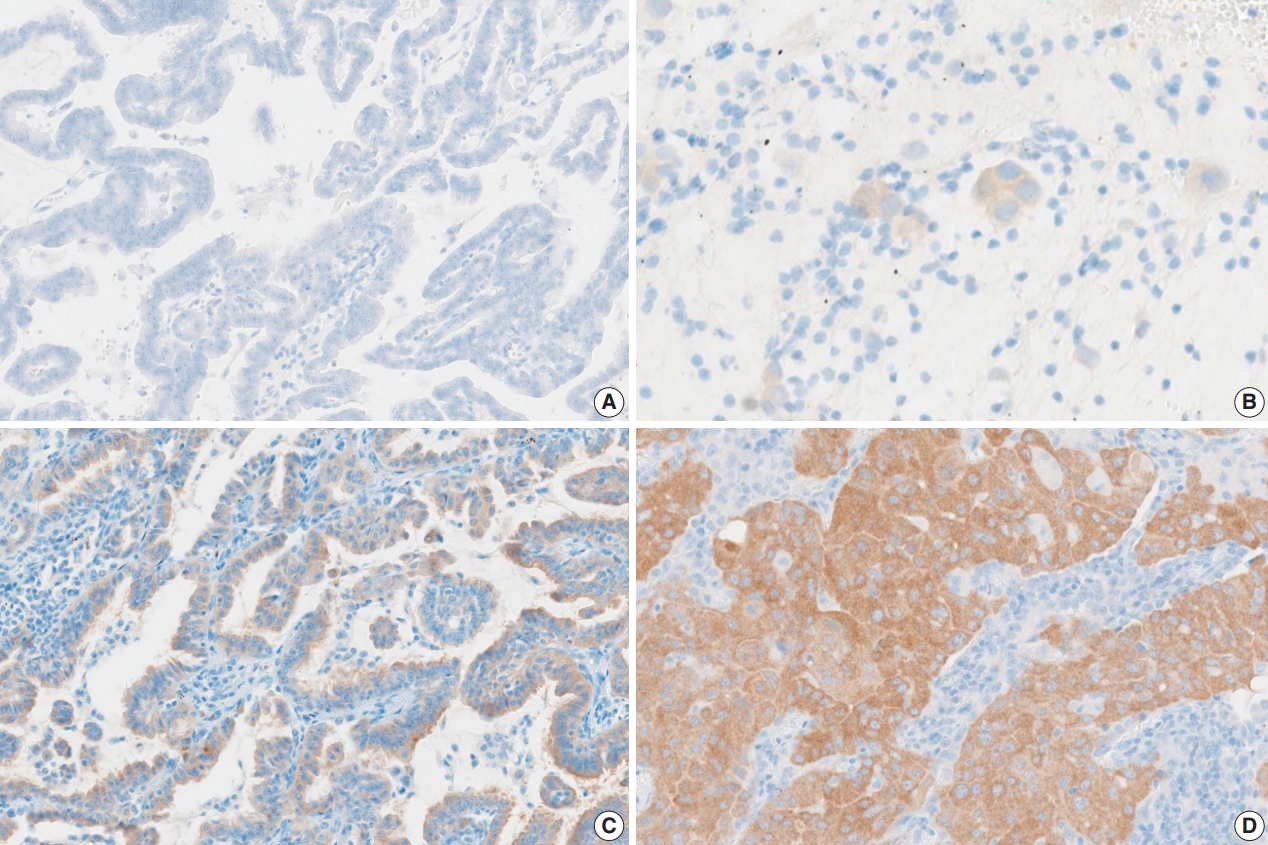
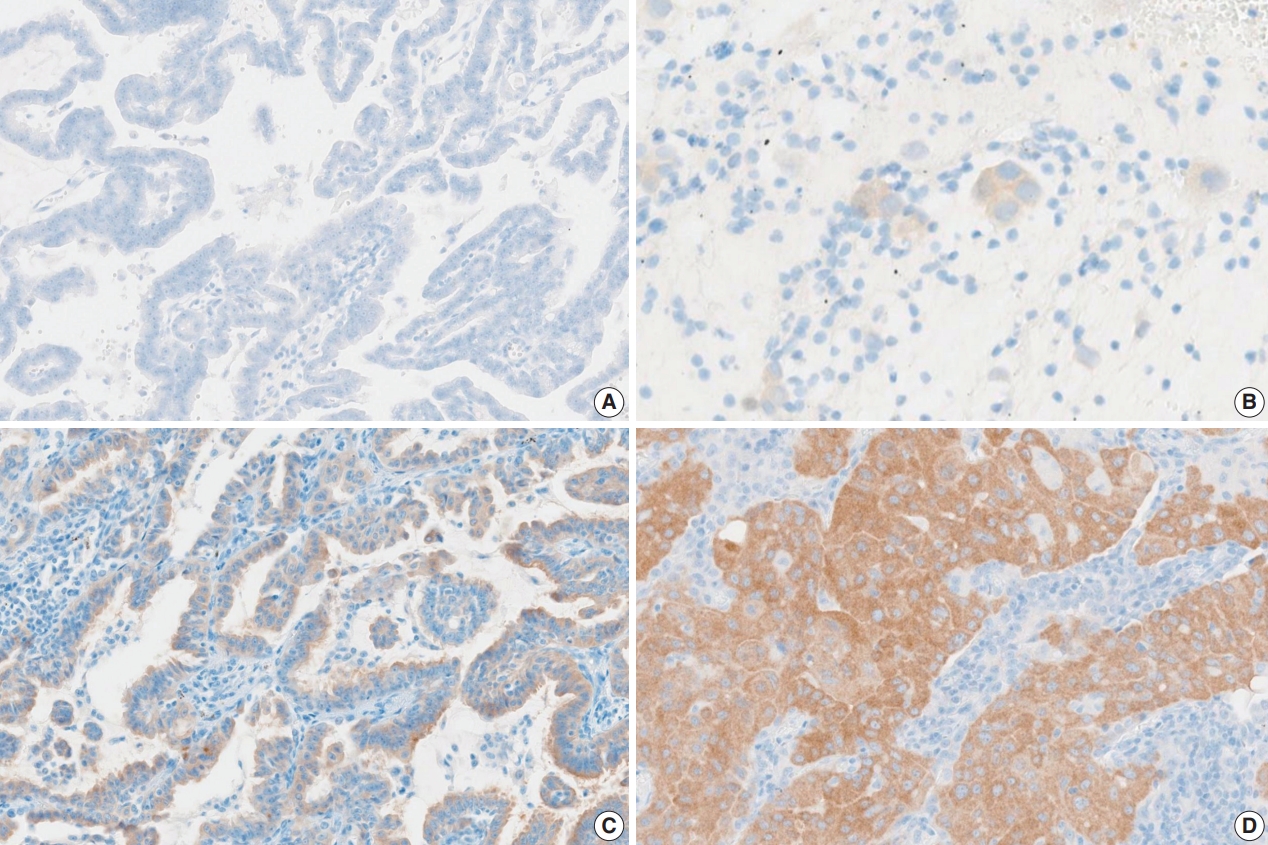
- Fifteen pathologists independently evaluated the whole slide images using Roche uPath enterprise software (Roche Diagnostics). Nine of 15 were pulmonary pathology specialists and six were surgical pathology fellows. Pathologists scored both the staining intensity and the percentage of tumor cell staining of any intensity (0%–100%, 5% increments). Tumor cell staining was defined as any perceptible cytoplasmic staining of viable tumor cells. The intensities were scored as “0” (negative staining), “1+” (weak staining), “2+” (moderate staining), and “3+” (strong staining) (Fig. 1).
- Statistical analysis
- Interobserver agreement for the percentage of tumor cell staining was evaluated by the intraclass correlation coefficients (ICCs). Interobserver agreement for staining intensity was evaluated by the Kendall concordance coefficient. ICC and Kendall concordance coefficient are interpreted as follows: <0.5 indicates poor agreement, between 0.5 and 0.75 indicates moderate agreement, between 0.75 and 0.9 indicates good agreement, and above 0.9 indicates excellent agreement. Differences in the concordance between the specialist group and fellow group were assessed by the Wilcoxon rank sum test.
- The receiver operating characteristic (ROC) curve was used to determine the cutoff value for the BRAF VE1 IHC analysis. ROC curves were analyzed based on the average percentage of tumor cell staining of 15 pathologists for each case. The cutoff value or more was interpreted as positive for BRAF VE1 IHC. The sensitivity, specificity, positive predictive value, and negative predictive value for detecting BRAF V600E mutation were calculated based on the consistent interpretation results of more than eight of 15 pathologists (Table 2). Interobserver agreement for the interpretation was evaluated by the Fleiss kappa coefficient. A kappa coefficient of <0.20 indicates poor, 0.21 to 0.40 indicates fair agreement, 0.41 to 0.60 indicates moderate agreement, 0.61 to 0.80 indicates substantial agreement, and greater than 0.80 indicates almost perfect agreement. Statistical analysis was performed with SPSS ver. 25 (IBM Corp., Armonk, NY, USA).
MATERIALS AND METHODS
- Interobserver agreement for IHC scoring
- In the entire study group (n=30), the percentage of tumor cell staining and staining intensity showed good interobserver agreements (percentage of tumor cell staining, ICC=0.878 [95% confidence interval (CI), 0.813 to 0.930]; staining intensity, kappa=0.804). The results in the lung adenocarcinoma subset (n=20) also displayed good interobserver agreements for the percentage of tumor cell staining and staining intensity (percentage of tumor cell staining, ICC=0.869 [95% CI, 0.786 to 0.935]; staining intensity, kappa=0.849). Only the staining intensity exhibited a discrepancy between specialists and fellows (p=0.029). VE1 staining tended to be weaker and more heterogeneous in lung adenocarcinomas than PTCs and colon adenocarcinomas.
- Diagnostic performance of BRAF VE1 IHC
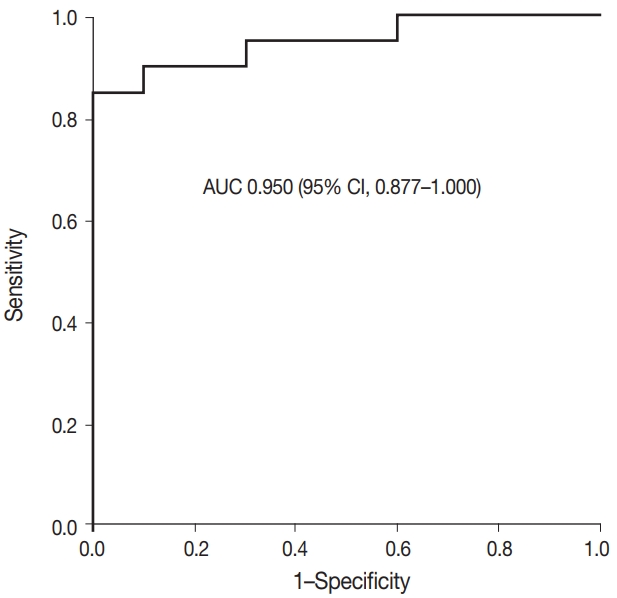
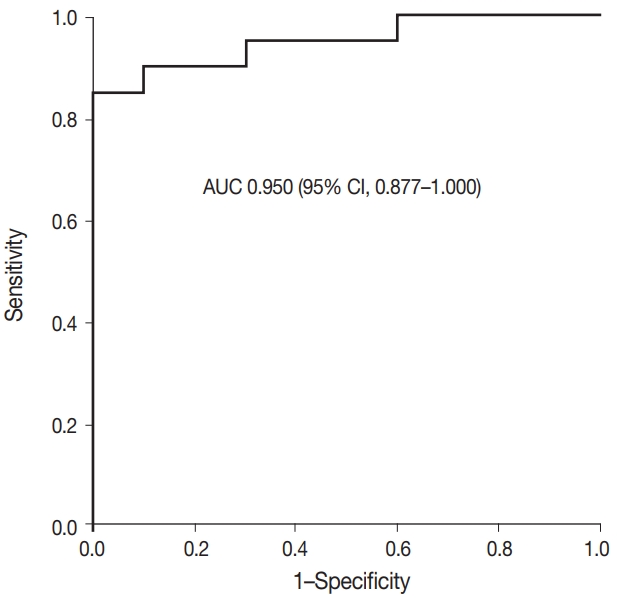
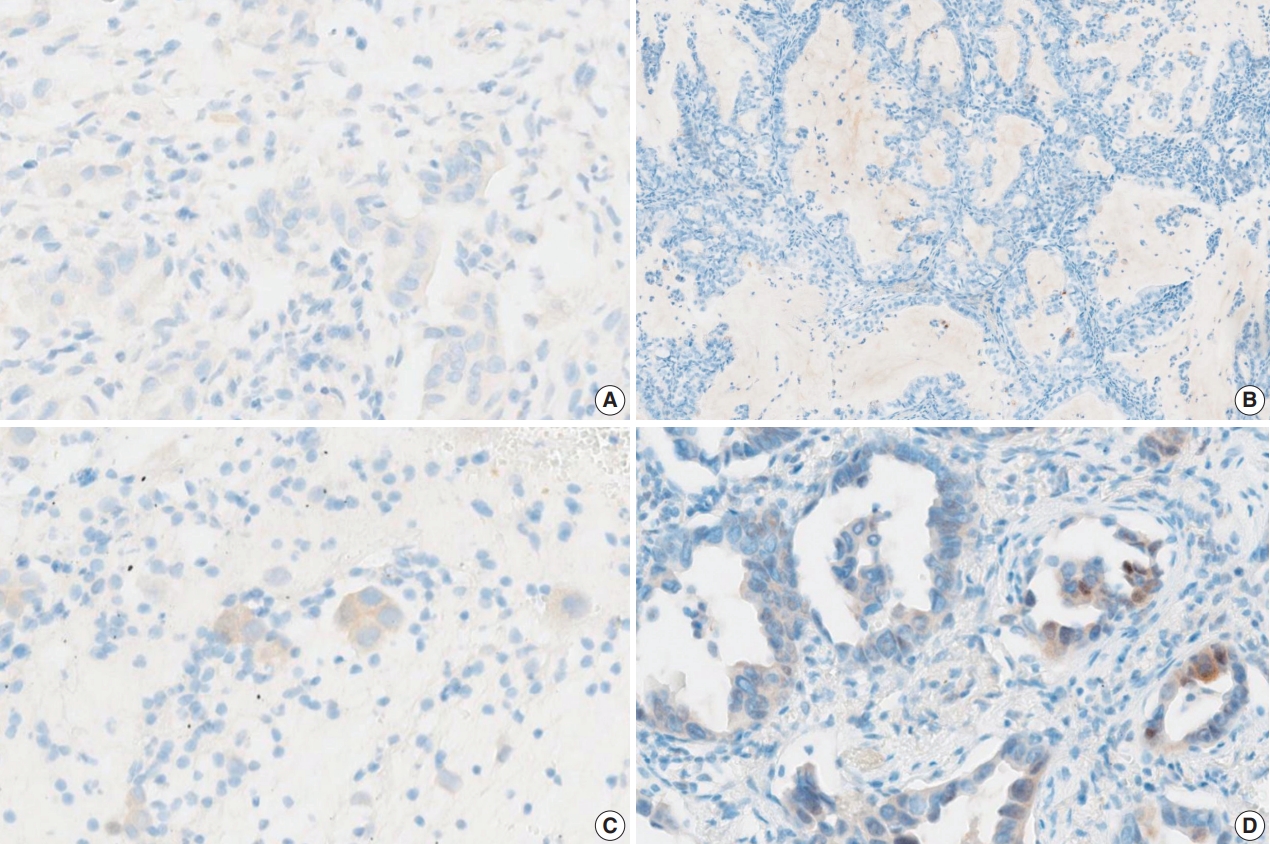
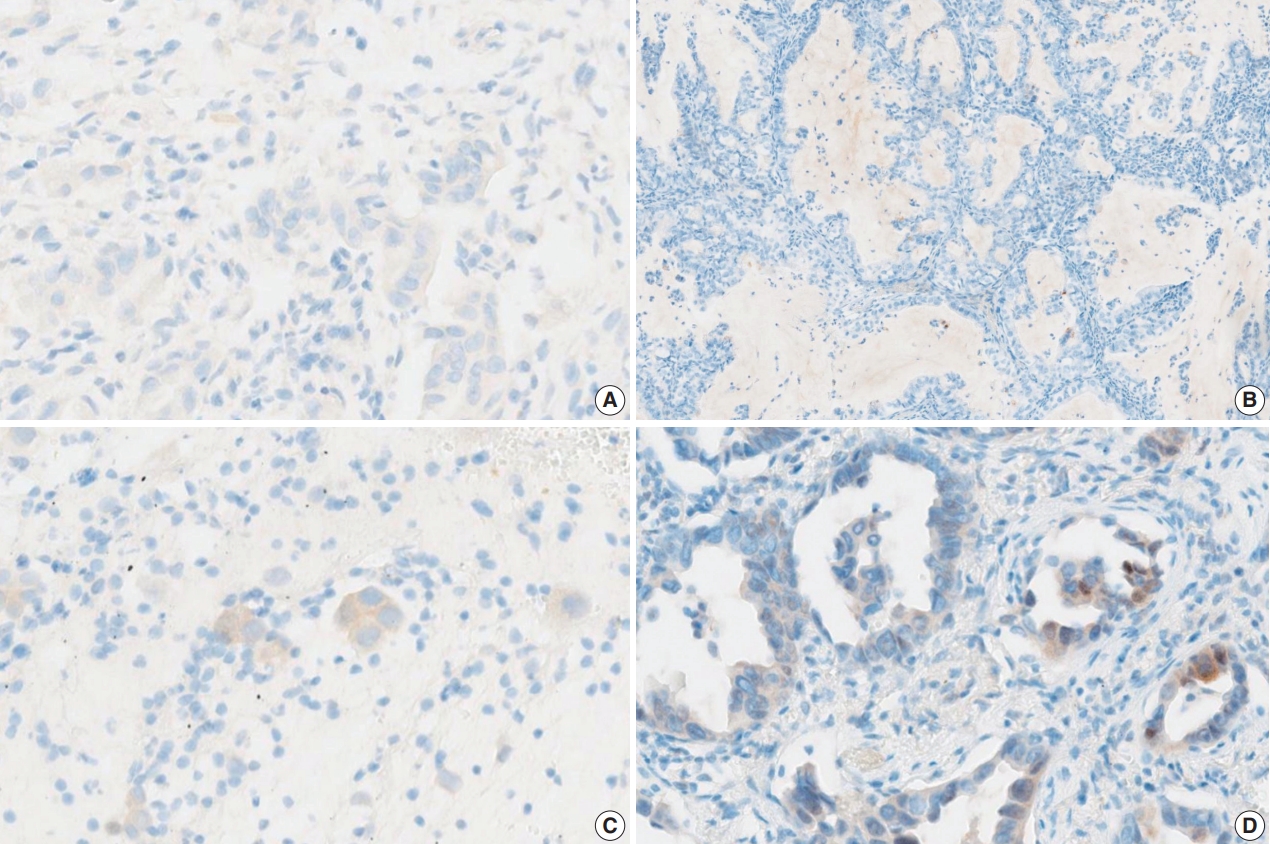
- Fig. 2 shows the ROC curve for the estimated diagnostic performance of the percentage of tumor cell staining in detecting BRAF V600E mutation. The area under the ROC curve was 0.950 (95% CI, 0.877 to 1.000). The 42.5% cutoff value maximized both sensitivity and specificity for the BRAF V600E mutation. Since the percentage of tumor cell staining was measured in increments of 5%, the cutoff value positive for BRAF VE1 IHC was defined as 40%. When 40% or more was considered positive for BRAF VE1 IHC, the sensitivity, specificity, positive predictive value, and negative predictive value of BRAF VE1 IHC were 90.0%, 90.0%, 94.7%, and 81.8%, respectively, for the entire study group. For the lung adenocarcinoma subset, sensitivity, specificity, positive predictive value, and negative predictive value of BRAF VE1 IHC were 80.0%, 90.0%, 88.9%, and 81.8%, respectively. False-negative results for BRAF VE1 IHC were shown in two resected lung adenocarcinoma cases (cases 7 and 14) (Table 2). Case 7 was interpreted as negative by 10 of 15 pathologists (Fig. 3A). Case 14, interpreted as negative by all pathologists, showed weak cytoplasmic staining in less than 5% of tumor cells (Fig. 3B). False-positive result for BRAF VE1 IHC was shown in one EBUS-guided biopsy specimen (case 22), which was interpreted as positive by 14 of 15 pathologists. Case 22 showed weak but diffuse cytoplasmic staining, but it turned out to be negative for the BRAF V600E mutation test (Fig. 3C).
- The interobserver agreement for the interpretation using the cutoff of 40% was almost perfect in the entire study group and the lung adenocarcinoma subset (entire study group, kappa=0.845 [95% CI, 0.810 to 0.880]; lung adenocarcinoma subset, kappa= 0.815 [95% CI, 0.772 to 0.858]). The interpretations of all 15 observers were consistent in 22 of 30 cases (73%), which includes 16 of 20 BRAF V600E–mutant cases (80%) and six of 10 BRAF V600E-negative cases (60%). Among the eight discrepant cases, cases 7 and 24 showed the greatest interobserver discrepancy (Table 2). Case 7 showed very faintly, questionable cytoplasmic staining. Thus, the percentage of tumor cell staining varied from 0% to 90% (mean 24%, standard deviation 34), depending on whether the observer considered the faint staining to be significant (Fig. 3A). Case 24, interpreted as negative by nine of 15 pathologists, showed heterogeneous weak to moderate staining within individual glands (Fig. 3D). Thus, the percentage of tumor cell staining was varied from 5% to 50% (mean 25%, standard deviation 20). In case 6, interpreted as positive by 12 of 15 pathologists, the percentage of tumor cell staining varied from 30% to 90% (mean±standard deviation, 56%±20%) due to heterogenous zonal staining with weak to moderate intensity. The other five cases showed discrepancies in one or two observers.
- Considering the presence of moderate to strong granular cytoplasmic staining in any tumor cells as positive criteria, sensitivity, specificity, positive predictive value, and negative predictive value were 85.0%, 90.0%, 94.4%, and 75.0% in the entire study group and 70.0%, 90.0%, 87.5%, and 75.0% in the lung adenocarcinomas. Complete agreement for all observers was obtained in four cases of 10 non-BRAF V600E mutant cases (40%) and eight of 20 BRAF V600E cases including one false-negative case (40%).
RESULTS
- This study shows that interobserver agreement for BRAF VE1 IHC interpretation was almost perfect (kappa=0.815) in lung adenocarcinoma, similar to the results of previous studies. Karbel et al. [22] reported almost perfect agreement (kappa=1.0) for BRAF VE1 IHC interpretation with three pathologists in 53 lung cancers. Previous studies on the interobserver agreement for BRAF VE1 IHC interpretation in PTCs, melanomas, and CRCs reported moderate to perfect agreement (kappa=0.554–1.0) [12-20]. But, in most studies, two or three pathologists interpreted the results of IHC. The study by Marin et al. [14] involved the largest number of pathologists and showed almost perfect agreement (kappa=0.81) for seven pathologists in 67 cases of melanoma. Fifteen pathologists were involved in this study, which is the largest number of pathologists to our knowledge. This study also evaluated interobserver agreement for the percentage of tumor cell staining and staining intensity and showed good agreement. The interobserver disagreement resulted from discrepancies in the interpretation of heterogeneous staining patterns and tumor cells showing weak staining intensity.
- In this study, the sensitivity and specificity of BRAF VE1 IHC were 80.0% and 90.0% in the lung adenocarcinomas, which are slightly lower than those reported in other studies on lung cancer (Table 1) [7,8,21,22]. This is probably because our study had a smaller sample size with a variety of types of specimens such as resection, biopsy, and EBUS specimens.
- The positive criteria for BRAF VE1 IHC have not yet been established. This study used a 40% cutoff for the interpretation of BRAF VE1 IHC irrespective of staining intensity according to the ROC curve. Previous studies reported false-positives in cases with heterogeneous non-diffuse cytoplasmic staining of variable intensity [3,14,16]. Dvorak et al. [3] recommended that cases showing heterogeneous cytoplasmic staining should be interpreted with caution. In melanomas, the unequivocal (≥1+) cytoplasmic staining of most tumor cells was used as positive criteria [10]. In lung cancers, Ilie et al. [6] used strong, homogenous staining as positive criteria. Hofman et al. [21] used at least 80% of tumor cells showing strong and homogenous staining as positive criteria, similar to Ilie et al. [6]. Our wild-type BRAF/VE1 positive case showed weak (1+) but diffuse cytoplasmic staining without nuclear staining. Nevertheless, if weak-stained cases are considered to be negative, the sensitivity decreases from 80.0% to 70.0% in lung adenocarcinoma. In consideration of the role of BRAF VE1 IHC as a screening test, the case with weak but diffuse cytoplasmic staining on BRAF VE1 IHC should be considered to be positive, and recommend further molecular tests for BRAF V600E mutation [3,16]. Sasaki et al. [5], Gow et al. [7], and Karbel et al. [22] used at least 50% of tumor cells with positive staining irrespective of staining intensity as positive criteria in lung cancers. In this study, even when the cutoff value was increased from 40% to 50%, the overall sensitivity and specificity were the same in lung adenocarcinoma. But, sensitivity to each pathologist decreased slightly in four out of 15 pathologists. Hwang et al. [8] interpreted cases showing at least weak and focal staining as positive. In this study, If Hwang’s criteria [8] are adopted, the sensitivity increases from 80% to 90%, but the specificity decreases from 90% to 50%. Further studies to validate the positive cutoff and/or staining intensity of BRAF VE1 IHC in the sensitive prediction of BRAF V600E mutation will be required.
- Various pre-analytical and analytical factors may affect the sensitivity and specificity of BRAF VE1 IHC. It is controversial whether there is a difference in sensitivity depending on the platform [3,10,23]. The Ventana platform can produce more optimal staining than the Dako or Leica platforms [16,23-25]. However, Sasaki et al. [5] (Dako platform) and Karbel et al. [22] (BioSB platform) reported similar results to other studies using Ventana platforms in lung cancers (Table 1). Antigen retrieval methods may affect the sensitivity of IHC [3,23]. Acidic buffers such as citrate buffer or Bond Epitope Retrieval Solution 1 (pH 6) may result in suboptimal staining [23,26,27]. Tris or EDTA buffers (pH 8) were recommended for retrieval agents [3,23]. As reported by Hwang et al. [8] in this study, lung adenocarcinomas showed more heterogeneous staining patterns and staining intensity than PTC and colon adenocarcinomas. The NordiQC data for BRAF VE1 IHC recommends the OptiView amplification kit–based protocol based for optimal results [23]. OptiView amplification kits, used to improve the visualization of ALK D5F3 IHC, could be also helpful in BRAF VE1 IHC in NSCLC. Rigorous antibody validation and protocol optimization, and quality control are required for the clinical application of BRAF VE1 IHC [9,16-18].
- The cross-reactivity of the VE1 antibody to non-V600E mutations, different BRAF point mutations, or unknown epitopes may cause false-negative results. In lung cancer, VE1 positivity has been reported in two of 72 cases with non-V600E mutations across previous studies to date [4-8]. Low tumor cell content may be attributed to false positivity [4,7,14]. Our false-positive case (case 22) was an EBUS biopsy sample with adequate tumor purity (20%). However, the total trimming-out of tumor cell contents from the sections for molecular tests is possible in a small biopsy sample and may affect the false-positive results. Decalcifying agents containing strong acids can affect DNA yield and protein expression and cause false-positive or false-negative results [1]. Cryofixation may cause false-negative results due to low expression of target proteins from tissue damage [10,11,28]. Our false-positive or -negative cases did not undergo decalcification or cryofixation. Signet ring cell morphology has been reported as common pitfall of VE1 interpretation in CRCs [16,17]. Signet ring cells can cause false-negative results due to minimal cytoplasm, which was almost entirely replaced by mucin [17]. On the other hand, the VE1 antibody could cross-react with intracellular mucin and result in false positivity [18]. This study had no signet ring cell component in false-positive or false-negative cases.
- The limitation of our study is the relatively small sample size and potential selection bias. Nevertheless, together with previous reports, this study suggests that BRAF VE1 IHC could be a screening test for further molecular testing of BRAF V600E mutation.
- BRAF VE1 IHC could be a screening test for the detection of BRAF V600E mutation in NSCLC. However, to introduce BRAF VE1 IHC into clinical practice, further studies are needed to optimize the protocol and to establish and validate interpretation criteria for BRAF VE1 IHC.
DISCUSSION
Ethics Statement
This study was approved by the Institutional Review Board of Inje University Ilsan Paik Hospital with a waiver of informed consent (2022-01-002).
Availability of Data and Material
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
Author contributions
Conceptualization: GKL, SYH. Data curation: SC. Formal analysis: SC. Investigation: YLC, GKL, SYH. Methodology: SC, YLC, HSS. Project administration: GKL, SYH. Supervision: GKL, SYH. Writing—original draft preparation: SC. Writing—review & editing: YLC, HSS, GKL, SYH. Approval of final manuscript: all authors.
Conflicts of Interest
The authors declare that they have no potential conflicts of interest.
Funding Statement
No funding to declare.
Acknowledgments
| Study | No. of cases | Cases with BRAF V600E mutation | Manufacturer | Platform | Molecular testing | Positive criteria | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|---|
| Sasaki et al. [5] | 26a | 5 | Dako | EnVision FLEX system | RT-PCR | ≥ 50% of tumor cells, any intensity | 100 | 95.2 |
| Ilie et al. [6] | 450b | 21 | Ventana | BenchMark XT | Direct sequencing, pyrosequencing | All tumor cells, strong and homogenous staining | 90.5 | 100 |
| Gow et al. [7] | 99a | 29 | Ventana | Benchmark XT | Direct sequencing, RT-PCR | ≥ 50% of tumor cells, any intensity | 96.55 | 98.57 |
| Karbel et al. [22] | 53c | 5 | Bio SB | PolyDetector Detection Systems | SSCP-PCR | ≥ 50% of tumor cells, any intensity | 97.9 | 100 |
| Seto et al. [4] | 219d | 14 | Ventana | Benchmark XT | Luminex GENOSEARCH BRAF, RT-PCR | N/A | 92.9 | 100 |
| Hofman et al. [21] | 1,317c | 32 | Ventana | Benchmark ULTRA | NGS, pyrosequencing | ≥ 80% of tumor cells, strong and homogenous staining | 100 | 100 |
| Hwang et al. [8] | 39e | 20 | Ventana | Benchmark ULTRA | NGS | At least weak and focal staining | 90.0 | 92.3 |
RT-PCR, reverse transcription polymerase chain reaction; SSCP, single-stranded conformation polymorphism; N/A, not available; NGS, next-generation sequencing.
aAdenocarcinomas;
bEGFR, KRAS, PI3KCA, Her2, and ALK wild-type non–small cell lung cancers (NSCLCs);
cNSCLCs;
d218 NSCLC cases and one small cell lung cancer case;
eConfirmed BRAF-mutated NSCLCs by NGS.
|
Case No. |
||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Negative (non-V600E) |
Positive (V600E) |
|||||||||||||||||||||||||||||
|
Lung ADC |
Lung ADC |
PTC |
Colon ADC |
|||||||||||||||||||||||||||
| 8 | 11 | 15 | 20 | 26 | 28 | 17 | 4a | 24 | 22b | 14 | 7c | 6 | 1 | 5b | 9 | 12 | 19 | 29 | 30b | 10 | 13 | 21 | 16 | 2 | 3 | 23 | 25 | 27 | 18 | |
| IHC | N | N | N | N | N | N | N | N | N | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| Specialist | ||||||||||||||||||||||||||||||
| 1 | N | N | N | N | N | N | N | N | N | P | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 2 | N | N | N | N | N | N | N | N | P | P | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 3 | N | N | N | N | N | N | N | N | P | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 4 | N | N | N | N | N | N | N | N | P | P | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 5 | N | N | N | N | N | N | N | N | N | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 6 | N | N | N | N | N | N | N | N | P | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 7 | N | N | N | N | N | N | N | N | N | P | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 8 | N | N | N | N | N | N | N | P | P | P | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 9 | N | N | N | N | N | N | N | N | N | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| Fellow | ||||||||||||||||||||||||||||||
| 1 | N | N | N | N | N | N | N | N | N | P | N | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 2 | N | N | N | N | N | N | N | N | N | N | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 3 | N | N | N | N | N | N | P | N | P | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 4 | N | N | N | N | N | N | N | P | N | P | N | N | N | P | P | P | P | P | P | P | P | P | P | N | P | P | P | P | P | N |
| 5 | N | N | N | N | N | N | N | N | N | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 6 | N | N | N | N | N | N | N | N | N | P | N | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
- 1. Chang S, Shim HS, Kim TJ, et al. Molecular biomarker testing for non-small cell lung cancer: consensus statement of the Korean Cardiopulmonary Pathology Study Group. J Pathol Transl Med 2021; 55: 181-91. ArticlePubMedPMCPDF
- 2. Sepulveda AR, Hamilton SR, Allegra CJ, et al. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. Arch Pathol Lab Med 2017; 141: 625-57. ArticlePubMedPDF
- 3. Dvorak K, Higgins A, Palting J, Cohen M, Brunhoeber P. Immunohistochemistry with anti-BRAF V600E (VE1) mouse monoclonal antibody is a sensitive method for detection of the BRAF V600E mutation in colon cancer: evaluation of 120 cases with and without KRAS mutation and literature review. Pathol Oncol Res 2019; 25: 349-59. ArticlePubMedPMCPDF
- 4. Seto K, Haneda M, Masago K, et al. Negative reactions of BRAF mutation-specific immunohistochemistry to non-V600E mutations of BRAF. Pathol Int 2020; 70: 253-61. ArticlePubMedPDF
- 5. Sasaki H, Shimizu S, Tani Y, et al. Usefulness of immunohistochemistry for the detection of the BRAF V600E mutation in Japanese lung adenocarcinoma. Lung Cancer 2013; 82: 51-4. ArticlePubMed
- 6. Ilie M, Long E, Hofman V, et al. Diagnostic value of immunohistochemistry for the detection of the BRAFV600E mutation in primary lung adenocarcinoma Caucasian patients. Ann Oncol 2013; 24: 742-8. PubMed
- 7. Gow CH, Hsieh MS, Lin YT, Liu YN, Shih JY. Validation of immunohistochemistry for the detection of BRAF V600E-mutated lung adenocarcinomas. Cancers (Basel) 2019; 11: 886.ArticlePubMedPMC
- 8. Hwang I, Choi YL, Lee H, et al. Selection strategies and practical application of BRAF V600E-mutated non-small cell lung carcinoma. Cancer Res Treat 2022; 54: 782-92. ArticlePubMedPMCPDF
- 9. O’Brien O, Lyons T, Murphy S, Feeley L, Power D, Heffron C. BRAF V600 mutation detection in melanoma: a comparison of two laboratory testing methods. J Clin Pathol 2017; 70: 935-40. ArticlePubMed
- 10. Anwar MA, Murad F, Dawson E, Abd Elmageed ZY, Tsumagari K, Kandil E. Immunohistochemistry as a reliable method for detection of BRAF-V600E mutation in melanoma: a systematic review and meta-analysis of current published literature. J Surg Res 2016; 203: 407-15. ArticlePubMed
- 11. Pyo JS, Sohn JH, Kang G. BRAF immunohistochemistry using clone VE1 is strongly concordant with BRAF(V600E) mutation test in papillary thyroid carcinoma. Endocr Pathol 2015; 26: 211-7. ArticlePubMedPDF
- 12. Schafroth C, Galvan JA, Centeno I, et al. VE1 immunohistochemistry predicts BRAF V600E mutation status and clinical outcome in colorectal cancer. Oncotarget 2015; 6: 41453-63. ArticlePubMedPMC
- 13. Rusu S, Verocq C, Trepant AL, et al. Immunohistochemistry as an accurate tool for the assessment of BRAF V600E and TP53 mutations in primary and metastatic melanoma. Mol Clin Oncol 2021; 15: 270.ArticlePubMedPMC
- 14. Marin C, Beauchet A, Capper D, et al. Detection of BRAF p.V600E mutations in melanoma by immunohistochemistry has a good interobserver reproducibility. Arch Pathol Lab Med 2014; 138: 71-5. ArticlePubMed
- 15. Fisher KE, Cohen C, Siddiqui MT, Palma JF, Lipford EH, Longshore JW. Accurate detection of BRAF p.V600E mutations in challenging melanoma specimens requires stringent immunohistochemistry scoring criteria or sensitive molecular assays. Hum Pathol 2014; 45: 2281-93. ArticlePubMed
- 16. Bledsoe JR, Kamionek M, Mino-Kenudson M. BRAF V600E immunohistochemistry is reliable in primary and metastatic colorectal carcinoma regardless of treatment status and shows high intratumoral homogeneity. Am J Surg Pathol 2014; 38: 1418-28. ArticlePubMedPMC
- 17. Toon CW, Walsh MD, Chou A, et al. BRAFV600E immunohistochemistry facilitates universal screening of colorectal cancers for Lynch syndrome. Am J Surg Pathol 2013; 37: 1592-602. ArticlePubMedPMC
- 18. Rossle M, Sigg M, Ruschoff JH, et al. Ultra-deep sequencing confirms immunohistochemistry as a highly sensitive and specific method for detecting BRAF V600E mutations in colorectal carcinoma. Virchows Arch 2013; 463: 623-31. ArticlePubMedPDF
- 19. Capper D, Voigt A, Bozukova G, et al. BRAF V600E-specific immunohistochemistry for the exclusion of Lynch syndrome in MSI-H colorectal cancer. Int J Cancer 2013; 133: 1624-30. ArticlePubMed
- 20. Affolter K, Samowitz W, Tripp S, Bronner MP. BRAF V600E mutation detection by immunohistochemistry in colorectal carcinoma. Genes Chromosomes Cancer 2013; 52: 748-52. ArticlePubMed
- 21. Hofman V, Benzaquen J, Heeke S, et al. Real-world assessment of the BRAF status in non-squamous cell lung carcinoma using VE1 immunohistochemistry: a single laboratory experience (LPCE, Nice, France). Lung Cancer 2020; 145: 58-62. ArticlePubMed
- 22. Karbel HA, Ejam SS, Naji AZ. Immunohistochemical study using monoclonal VE1 antibody can substitute the molecular tests for apprehension of BRAF V600E mutation in patients with non-smallcell lung carcinoma. Anal Cell Pathol (Amst) 2019; 2019: 2315673.ArticlePubMedPMCPDF
- 23. NordiQC. Assessment Run 62 2021 BRAF (BRAF V600E) [Internet]. Aalborg: NordiQC, 2021 [cited 2021 Nov 30]. Available from: https://www.nordiqc.org/downloads/assessments/146_113.pdf.
- 24. Sinicrope FA, Smyrk TC, Tougeron D, et al. Mutation-specific antibody detects mutant BRAFV600E protein expression in human colon carcinomas. Cancer 2013; 119: 2765-70. ArticlePubMedPMC
- 25. Adackapara CA, Sholl LM, Barletta JA, Hornick JL. Immunohistochemistry using the BRAF V600E mutation-specific monoclonal antibody VE1 is not a useful surrogate for genotyping in colorectal adenocarcinoma. Histopathology 2013; 63: 187-93. ArticlePubMed
- 26. Kuan SF, Navina S, Cressman KL, Pai RK. Immunohistochemical detection of BRAF V600E mutant protein using the VE1 antibody in colorectal carcinoma is highly concordant with molecular testing but requires rigorous antibody optimization. Hum Pathol 2014; 45: 464-72. ArticlePubMed
- 27. Lasota J, Kowalik A, Wasag B, et al. Detection of the BRAF V600E mutation in colon carcinoma: critical evaluation of the imunohistochemical approach. Am J Surg Pathol 2014; 38: 1235-41. PubMedPMC
- 28. Bullock M, O’Neill C, Chou A, et al. Utilization of a MAB for BRAF(V600E) detection in papillary thyroid carcinoma. Endocr Relat Cancer 2012; 19: 779-84. ArticlePubMed
REFERENCES
Figure & Data
References
Citations

- Dedifferentiated Leiomyosarcoma of the Uterine Corpus with Heterologous Component: Clinicopathological Analysis of Five Consecutive Cases from a Single Institution and Comprehensive Literature Review
Suyeon Kim, Hyunsik Bae, Hyun-Soo Kim
Diagnostics.2024; 14(2): 160. CrossRef - Differentiating BRAF V600E- and RAS-like alterations in encapsulated follicular patterned tumors through histologic features: a validation study
Chankyung Kim, Shipra Agarwal, Andrey Bychkov, Jen-Fan Hang, Agnes Stephanie Harahap, Mitsuyoshi Hirokawa, Kennichi Kakudo, Somboon Keelawat, Chih-Yi Liu, Zhiyan Liu, Truong Phan-Xuan Nguyen, Chanchal Rana, Huy Gia Vuong, Yun Zhu, Chan Kwon Jung
Virchows Archiv.2024; 484(4): 645. CrossRef - BRAF V600E Mutation of Non-Small Cell Lung Cancer in Korean Patients
Hyo Yeong Ahn, Chang Hun Lee, Min Ki Lee, Jung Seop Eom, Yeon Joo Jeong, Yeong Dae Kim, Jeong Su Cho, Jonggeun Lee, So Jeong Lee, Dong Hoon Shin, Ahrong Kim
Medicina.2023; 59(6): 1085. CrossRef - Reevaluating diagnostic categories and associated malignancy risks in thyroid core needle biopsy
Chan Kwon Jung
Journal of Pathology and Translational Medicine.2023; 57(4): 208. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure



Fig. 1.
Fig. 2.
Fig. 3.
| Study | No. of cases | Cases with BRAF V600E mutation | Manufacturer | Platform | Molecular testing | Positive criteria | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|---|
| Sasaki et al. [5] | 26 |
5 | Dako | EnVision FLEX system | RT-PCR | ≥ 50% of tumor cells, any intensity | 100 | 95.2 |
| Ilie et al. [6] | 450 |
21 | Ventana | BenchMark XT | Direct sequencing, pyrosequencing | All tumor cells, strong and homogenous staining | 90.5 | 100 |
| Gow et al. [7] | 99 |
29 | Ventana | Benchmark XT | Direct sequencing, RT-PCR | ≥ 50% of tumor cells, any intensity | 96.55 | 98.57 |
| Karbel et al. [22] | 53 |
5 | Bio SB | PolyDetector Detection Systems | SSCP-PCR | ≥ 50% of tumor cells, any intensity | 97.9 | 100 |
| Seto et al. [4] | 219 |
14 | Ventana | Benchmark XT | Luminex GENOSEARCH BRAF, RT-PCR | N/A | 92.9 | 100 |
| Hofman et al. [21] | 1,317 |
32 | Ventana | Benchmark ULTRA | NGS, pyrosequencing | ≥ 80% of tumor cells, strong and homogenous staining | 100 | 100 |
| Hwang et al. [8] | 39 |
20 | Ventana | Benchmark ULTRA | NGS | At least weak and focal staining | 90.0 | 92.3 |
| Case No. |
||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Negative (non-V600E) |
Positive (V600E) |
|||||||||||||||||||||||||||||
| Lung ADC |
Lung ADC |
PTC |
Colon ADC |
|||||||||||||||||||||||||||
| 8 | 11 | 15 | 20 | 26 | 28 | 17 | 4 |
24 | 22 |
14 | 7 |
6 | 1 | 5 |
9 | 12 | 19 | 29 | 30 |
10 | 13 | 21 | 16 | 2 | 3 | 23 | 25 | 27 | 18 | |
| IHC | N | N | N | N | N | N | N | N | N | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| Specialist | ||||||||||||||||||||||||||||||
| 1 | N | N | N | N | N | N | N | N | N | P | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 2 | N | N | N | N | N | N | N | N | P | P | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 3 | N | N | N | N | N | N | N | N | P | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 4 | N | N | N | N | N | N | N | N | P | P | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 5 | N | N | N | N | N | N | N | N | N | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 6 | N | N | N | N | N | N | N | N | P | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 7 | N | N | N | N | N | N | N | N | N | P | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 8 | N | N | N | N | N | N | N | P | P | P | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 9 | N | N | N | N | N | N | N | N | N | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| Fellow | ||||||||||||||||||||||||||||||
| 1 | N | N | N | N | N | N | N | N | N | P | N | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 2 | N | N | N | N | N | N | N | N | N | N | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 3 | N | N | N | N | N | N | P | N | P | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 4 | N | N | N | N | N | N | N | P | N | P | N | N | N | P | P | P | P | P | P | P | P | P | P | N | P | P | P | P | P | N |
| 5 | N | N | N | N | N | N | N | N | N | P | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
| 6 | N | N | N | N | N | N | N | N | N | P | N | N | N | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P | P |
RT-PCR, reverse transcription polymerase chain reaction; SSCP, single-stranded conformation polymorphism; N/A, not available; NGS, next-generation sequencing. Adenocarcinomas; EGFR, KRAS, PI3KCA, Her2, and ALK wild-type non–small cell lung cancers (NSCLCs); NSCLCs; 218 NSCLC cases and one small cell lung cancer case; Confirmed
IHC, immunohistochemistry; ADC, Adenocarcinoma; PTC, papillary thyroid carcinoma; N, negative; P, positive. Needle biopsy; Endobronchial ultrasound-guided biopsy; Bronchoscopic biopsy.

 E-submission
E-submission