Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 56(6); 2022 > Article
-
Review
The application of high-throughput proteomics in cytopathology -
Ilias P. Nikas1
 , Han Suk Ryu,2,3
, Han Suk Ryu,2,3
-
Journal of Pathology and Translational Medicine 2022;56(6):309-318.
DOI: https://doi.org/10.4132/jptm.2022.08.30
Published online: November 9, 2022
1School of Medicine, European University Cyprus, Nicosia, Cyprus
2Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
3Department of Pathology, Seoul National University Hospital, Seoul, Korea
- Corresponding Author: Han Suk Ryu, MD, PhD, Department of Pathology, Seoul National University College of Medicine, 103 Daehak-ro, Jongno-gu, Seoul 03080, Korea Tel: +82-2-740-8277, Fax: +82-2-743-5530, E-mail: nash77@snu.ac.kr
© 2022 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Figure & Data
References
Citations

- Identification of NIFTP-Specific mRNA Markers for Reliable Molecular Diagnosis of Thyroid Tumors
So-Yeon Lee, Jong-Lyul Park, Kwangsoon Kim, Ja Seong Bae, Jae-Yoon Kim, Seon-Young Kim, Chan Kwon Jung
Endocrine Pathology.2023; 34(3): 311. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure

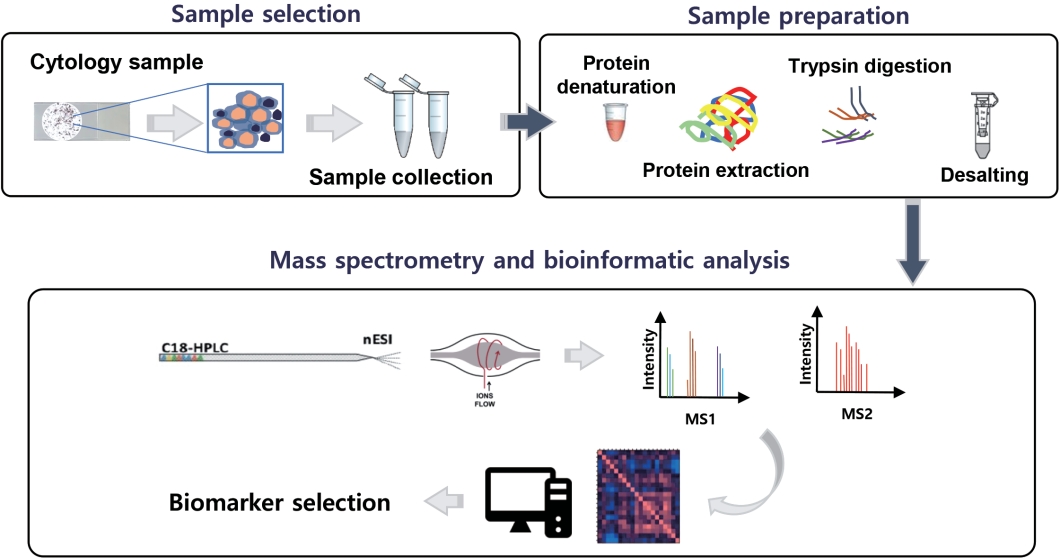
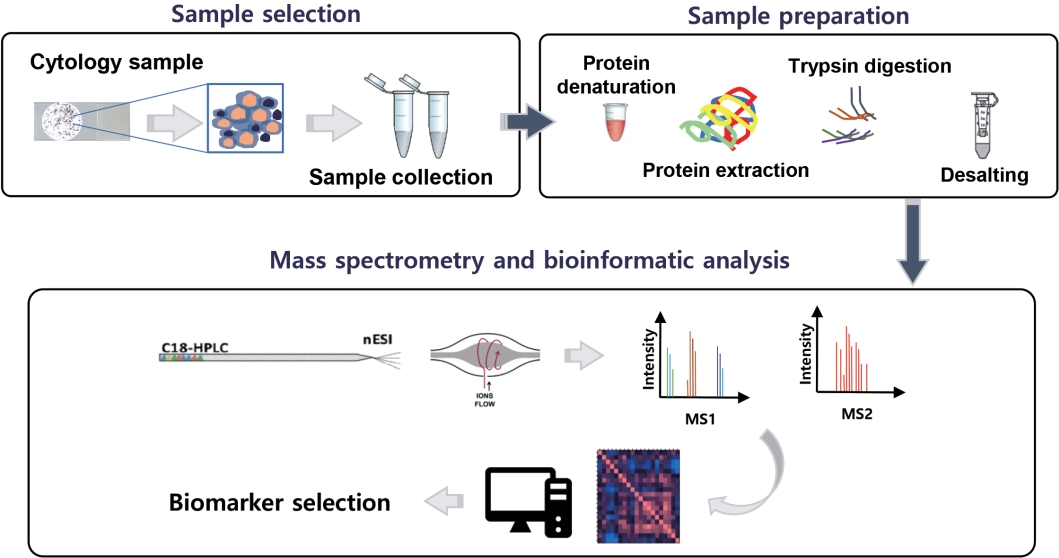
Fig. 1.
| Study | Sample type | No. of samples | High-throughput proteomics approach | Key findings | |
|---|---|---|---|---|---|
| Breast | |||||
| Pawlik et al. (2006) [62] | NAF | 18 from breast cancer (stages I and II); 4 controls | ICAT LC-MS/MS | Vitamin D binding protein precursor was overexpressed in the NAF of patients with early-stage breast cancer compared to controls | |
| Pawlik et al. (2005) [42] | NAF | 23 from breast cancer (stages I and II); 5 controls | SELDI-MS | Significant proteomic profile differences were found in the NAF of patients with early-stage breast cancer compared to controls | |
| Sauter et al. (2005) [39] | NAF | 27 from breast cancer; 87 controls | SELDI-MS | Proteomic profile differences were found in the NAF of patients with DCIS compared to controls, and invasive cancer compared to DCIS | |
| Alexander et al. (2004) [33] | NAF | 52 from DCIS and invasive cancer; 53 controls | 2D PAGE and MALDI-MS | GCDFP-15 was significantly underexpressed and AAG overexpressed in the breast cancer samples tested | |
| Sauter et al. (2002) [43] | NAF | 20 from breast cancer; 13 controls | SELDI-MS | Proteomic profile differences (5 proteins) were found in the NAF of patients with cancer compared to controls | |
| George et al. (2021) [44] | NAF | 9 from breast cancer; 4 controls | LC-MS/MS | Proteomic profile differences (40 proteins) were found in the NAF of patients with cancer compared to controls | |
| Pavlou et al. (2010) [63] | NAF | 3 from breast cancer; 3 controls | LC-MS/MS | More than 800 proteins were discovered, as part of the NAF proteome | |
| Noble et al. (2007) [64] | NAF | Paired samples from 21 patients with breast cancer; paired and unilateral samples from 44 controls | SELDI-MS | Whereas no proteomic profile differences were found in the NAF received from the breast with cancer compared to the contralateral healthy one, significant differences were identified between women with cancer (in both cancerous and healthy breasts) and healthy controls | |
| Fowler et al. (2004) [46] | FNA | 24 (benign and malignant lesions) | SELDI-MS | Liquid-based cytology samples, stored in the methanol-based PreservCyt, were suitable for satisfactory and reproducible proteomic analysis | |
| Franzen et al. (2019) [45] | FNA | 25 from breast cancer, 32 controls | PEA | Expression levels of several immune-related proteins differed between cancer and controls, while a few were associated with ER, Ki-67 status, and tumor grading | |
| Rapkiewicz et al. (2007) [47] | FNA | 63 (50 with cancer) from 21 patients | RPPM | The RPPM technology successfully identified and quantified selected proteins in FNA samples | |
| Thyroid | |||||
| Pagni et al. (2015) [48] | FNA | Samples from 6 patients (3 non-neoplastic, 1 Hurthle cell adenoma, 1 PTC, 1 MTC) | MALDI-MSI | Proteomic profile differences were identified between diverse thyroid lesions sampled with FNA | |
| Mainini et al. (2013) [49] | FNA | Samples from 7 patients (non-neoplastic and neoplastic) | MALDI-MSI | In situ proteomic analysis could differentiate between non-neoplastic and malignant lesions, identify PTC, also distinguish PTC cases carrying the BRAF V600E mutation | |
| Capitoli et al. (2020) [50] | FNA | Samples from 43 patients (non-neoplastic and neoplastic; training and validation cohorts) | MALDI-MSI | In situ proteomic analysis distinguished Hashimoto thyroiditis from hyperplastic nodules and PTC | |
| Pagni et al. (2016) [51] | FNA | 36 (13 benign, 10 indeterminate, 13 PTCs) | MALDI-MSI | In situ proteomic analysis distinguished benign thyroid lesions from PTCs and correctly triaged indeterminate FNA lesions as either benign or malignant | |
| Giusti et al. (2007) [65] | FNA | 17 suspicious and malignant thyroid lesions | 2D-GE and MALDI-MS | Several proteins were identified, involved in various cell processes (e.g., metabolism, apoptosis, motility) | |
| Giusti et al. (2008) [66] | FNA | 13 PTCs | 2D-GE and MALDI-MS | 17 proteins were overexpressed in thyroid cancer patients compared to controls; proteomic profile differences were also identified between classic and tall cell PTC variants | |
| Capitoli et al. (2022) [52] | FNA | 240 (internal and external validation cohorts) | MALDI-MSI | Whereas the diagnostic accuracy of the in situ proteomics-based classification model was inferior in the external than internal validation cohort, this was improved when sample cellularity was adequate | |
| Ciregia et al. (2016) [67] | FNA | 212 (benign, intermediate, suspicious for malignancy, and malignant) | 2D-GE and LC-ESI-MS/MS | Proteomic profile differences (25 proteins) were found between benign and malignant lesions; ROC curve analysis showed the combination of ENO1, ANXA1, DJ1, SOD, CRNN protein levels had the best discriminatory capacity | |
| Ucal et al. (2017) [68] | FNA | 18 (12 PTCs, 6 benign) | LC-MS/MS | Several actin cytoskeleton proteins (e.g., Arp 2/3 complex overexpression) were altered in PTC; IQGAP1 was upregulated in CV-PTC, while IQGAP2 in FV-PTC, at significant levels, respectively | |
| Capitoli et al. (2019) [53] | FNA | 28 (benign, intermediate, and malignant; training and validation cohorts) | MALDI-MSI | The in situ proteomics-based model was able to predict the classification derived from the FNA morphologic evaluation of the thyroid lesions | |
| Lin et al. (2019) [69] | FNA | 120 PTMCs (60 with LN metastasis, and 60 without) | TMT and LC-MS/MS | ISG15 levels distinguished PTMC patients developing LN metastasis from the ones that did not | |
| Urine | |||||
| Park et al. (2020) [70] | Urine (LBC cytology) | 16 (6 NIBUC, 5 SIBUC, and 5 MIBUC) | LC-MS/MS | Proteomic analysis of LBC samples revealed moesin as a biomarker predicting bladder urothelial cancer invasion | |
| Yang et al. (2011) [71] | Urine | 54 cancer, and 46 controls | LC-MS/MS | Overexpression of A1AT was associated with the presence of bladder urothelial cancer, at a significant level | |
| Theodorescu et al. (2006) [72] | Urine | 655 (non-malignant and malignant) | CE-MS | The model predicted the presence of urothelial cancer in urine samples with high diagnostic accuracy | |
| Lee et al. (2018) [73] | Urine (LBC cytology) | 20 (10 bladder cancer; 10 controls) | LC-MS/MS | Proteomic analysis revealed AHNAK as a biomarker differentiating bladder cancer from controls in LBC cytology samples | |
| Pap test | |||||
| Schwamborn et al. (2011) [54] | Pap test | 32 (18 with LSIL or higher; 14 NILM) | MALDI-MSI | In situ proteomics analysis was able to correctly assign most lesions into their original cytologic classification group | |
| Boylan et al. (2014) [74] | Pap test | 100, all with normal cytology | 1D PAGE and LC-MS/MS | The core proteome of normal Pap test, comprising 153 proteins, was created by proteomics analysis of residual LBC samples | |
| Boylan et al. (2021) [75] | Pap test | One patient with serous ovarian cancer | LC/MS/MS | LBC is suitable for high-throughput proteomic analysis to identify ovarian cancer biomarkers | |
| Effusions | |||||
| Schwamborn et al. (2019) [55] | Pleural and peritoneal effusions | 24 with serous ovarian cancer, 19 with non-ovarian cancers | MALDI-MSI | In situ proteomic analysis was able to differentiate among diverse cancer types in effusions | |
| Perzanowska et al. (2018) [56] | Pleural effusion | 69 malignant, 49 benign (controls) | LC/MRM-MS | Multiplex proteomic analysis was able to differentiate between benign and malignant effusions, besides among lung cancer histologic subtypes (SCC, AC, SqCC) | |
| Li et al. (2016) [57] | Pleural effusion | 83 malignant (lung ACs), 60 benign (training and validation cohorts) | MALDI-MS | The model was able to differentiate between benign and malignant effusions with high diagnostic accuracy; CARD9 was downregulated in malignant effusions | |
| Liu et al. (2015) [76] | Pleural effusion | 405 malignant and benign effusions (discovery and validation cohorts) | 1D-PAGE and LC-MS/MS | Overexpression of MET, DPP4, and PTPRF identified metastatic lung adenocarcinomas in effusion samples with high diagnostic accuracy | |
| Li et al. (2015) [77] | Pleural effusion | 6 (3 NSCLC, 3 TB) | 1D-PAGE and LC/MS/MS | Proteomic analysis was able to differentiate NSCLC from TB effusions; IL1A was overexpressed in NSCLC compared to TB effusions | |
| Hegmans et al. (2009) [78] | Pleural effusion | 89 (mesothelioma, metastatic carcinoma, benign effusions) | SELDI-MS | SMRP was identified as a diagnostic biomarker of mesothelioma in pleural effusions | |
| Pancreatobiliary | |||||
| Inoue et al. (2022) [58] | EUS-FNA | 40 PDAC, 6 AIP | LC-MS/MS | Expression of several EV proteins differed between PDAC and AIP patients | |
| Lee et al. (2012) [59] | EUS-FNA | 5 BD-IPMNs, 5 inflammatory cysts | Cytokine microarray | HGF and GM-CSF differentiated inflammatory cysts from BD- IPMNs | |
| Navaneethan et al. (2015) [60] | Bile | 24 (PDAC, CCA, PSC, other non-neoplastic) | SDS-PAGE and LC-MS/MS | Expression of several proteins differed between malignant and non- neoplastic biliary strictures | |
| Salivary | |||||
| Seccia et al. (2020) [61] | FNA | 20 MSGTs, 37 PAs, 14 WTs | 2D-GE and LC-ESI-MS/MS | Overexpression of 4 proteins (annexin-5, cofilin-1, peptidyl-prolyl-cis–trans-isomerase-A, and F-actin-capping-alpha-1) differentiated MSGTs from benign aspirates | |
| Bone marrow | |||||
| Chen et al. (2021) [41] | Bone marrow aspirate | 5 RRMM, 5 NDMM | TMT-MS/MS | Overexpression of the biomarker SERPINB9 was found in RRMM, compared to NDMM | |
| Study | Cancer type/sample type | Novel biomarker(s) | Expression status in cancer |
|---|---|---|---|
| Pawlik et al. (2006) [62] | Breast/NAF | Vitamin D-binding protein precursor | Vitamin D-binding protein precursor: ↑ in breast cancer |
| Alexander et al. (2004) [33] | Breast/NAF | GCDFP-15, AAG | AAG: ↑ in breast cancer |
| GCDFP-15: ↓ in breast cancer | |||
| Ciregia et al. (2016) [67] | Thyroid/Thyroid FNA, serum, saliva | ANXA1 | ANXA1: ↑ in thyroid cancer |
| Ucal et al. (2017) [68] | Thyroid/FNA | IQGAP1, IQGAP2 | IQGAP1: ↑ in CV-PTC |
| IQGAP2: ↑ in FV-PTC | |||
| Lin et al. (2019) [69] | Thyroid/FNA | ISG15 | ISG15: ↑ in PTMC patients with metastasis to cervical lymph nodes (prognostic biomarker) |
| Giusti et al. (2008) [66] | Thyroid/FNA | TTR, FLC, proteasome activator complex subunit 1 and 2, alpha-1-antitrypsin precursor, GAPDH, LDH-B, Apo-A1, annexin A1, DJ-1 protein and cofilin-1 | TTR, FLC, proteasome activator complex subunit 1 and 2, alpha-1-antitrypsin precursor, GAPDH, LDH-B, Apo-A1, annexin A1, DJ-1 protein and cofilin-1: ↑ in PTC |
| Park et al. (2020) [70] | Bladder/Urine | Moesin | Moesin: ↑ in invasive bladder cancer |
| Yang et al. (2011) [71] | Bladder/Urine | A1AT | A1AT: ↑ in bladder cancer |
| Lee et al. (2018) [73] | Bladder/Urine | AHNAK | AHNAK: ↑ in bladder cancer |
| Li et al. (2016) [57] | Lung/Effusions | CARD9 | CARD9: ↓ in malignant effusions |
| Liu et al. (2015) [76] | Lung/Effusions | MET, DPP4, and PTPRF | MET, DPP4, and PTPRF: ↑ in malignant effusions |
| Li et al. (2015) [77] | Lung/Effusions | IL1A | IL1A: ↑ in malignant effusions |
| Hegmans et al. (2009) [78] | Mesothelioma/Effusions | SMRP | SMRP: ↑ in mesothelioma |
| Seccia et al. (2020) [61] | MSGTs/FNA | Annexin-5, cofilin-1, peptidyl-prolyl-cis–trans-isomerase-A and F-actin-cappingalpha-1 | Annexin-5, cofilin-1, peptidyl-prolyl-cis–trans-isomerase-A and F-actin-capping-alpha-1: ↑ in MSGTs |
| Chen et al. (2021) [41] | MM/Bone marrow aspirate | SERPINB9 | SERPINB9: ↑ in RRMM (prognostic biomarker) |
NAF, nipple aspirate fluid; ICAT, isotope-coded affinity tag; LC-MS/MS, liquid chromatography-tandem mass spectrometry; SELDI-MS, surface-enhanced laser desorption/ionization-mass spectrometry; DCIS, ductal carcinoma in situ; 2D PAGE, two-dimensional polyacrylamide gel electrophoresis; MALDI, matrixassisted laser desorption ionization; GCDFP, gross cystic disease fluid protein; AAG, alpha1-acid glycoprotein; FNA, fine-needle aspiration; PEA, proximity extension assay; ER, estrogen receptor; RPPM, reverse-phase protein microarrays; PTC, papillary thyroid carcinoma; MTC, medullary thyroid carcinoma; MSI, mass spectrometry imaging; 2D-GE, two-dimensional gel electrophoresis; LC-ESI-MS/MS, liquid chromatography electrospray ionization tandem mass spectrometry; ROC, receiver operating characteristic; ENO1, enolase 1; ANXAI, annexin A1; DJ1, protein DJ-1; SOD, superoxide dismutase; CV-PTC, classic variant PTC; FV-PTC, follicular variant PTC; PTMC, papillary thyroid microcarcinoma; LN, lymph node; TMT, tandem mass tags; ISG15, interferon-stimulated gene 15 protein; LBC, liquid-based cytology; NIBUC, non-invasive bladder urothelial carcinoma; SIBUC, stromal-invasive bladder urothelial carcinoma; MIBUC, muscle-invasive bladder urothelial carcinoma; A1AT, alpha 1 antitrypsin; CE-MS, capillary electrophoresis coupled to mass spectrometry; LSIL, lowgrade squamous intraepithelial lesion; NILM, negative for intraepithelial lesion or malignancy; Pap, Papanicolaou; MRM, multiple reaction monitoring; SCC, small cell carcinoma; AC, adenocarcinoma; SqCC, squamous cell carcinoma; CARD9, caspase recruitment domain family member 9; DPP4, dipeptidyl peptidase-4; PTPRF, protein tyrosine phosphatase receptor type F; NSCLC, non–small cell lung cancer; TB, tuberculosis; SMRP, soluble mesothelin-related protein; EUS-FNA, endoscopic ultrasound-guided fine-needle aspiration; PDAC, pancreatic adenocarcinoma; AIP, autoimmune pancreatitis; EV, extracellular vesicles; BD-IPMNs, branch duct intraductal papillary mucinous neoplasms; HGF, hepatocyte growth factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; CCA, cholangiocarcinoma, PSC, primary sclerosing cholangitis; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; MSGTs, malignant salivary gland tumors; PAs, pleomorphic adenomas; WTs, Warthin tumors; RRMM, recurrent and relapsed multiple myeloma; NDMM, newly diagnosed multiple myeloma; TMT-MS, tandem mass tag-mass spectrometry.
NAF, nipple aspirate fluid; GCDFP, gross cystic disease fluid protein; AAG, alpha1-acid glycoprotein; FNA, fine-needle aspiration; ANXA1, annexin A1; IQGAP1, IQ motif containing GTPase activating protein 1; CV-PTC, classic variant PTC; FV-PTC, follicular variant PTC; ISG15, interferon-stimulated gene 15 protein; PTMC, papillary thyroid microcarcinoma; TTR, transthyretin; FLC, ferritin light chain; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; LDH-B, lactate dehydrogenase chain B; Apo-A1, apolipoprotein A1 precursor; A1AT, alpha 1 antitrypsin; CARD9, caspase recruitment domain family member 9; DPP4, dipeptidyl peptidase-4; PTPRF, protein tyrosine phosphatase receptor type F; IL1A, interleukin 1A; SMRP, soluble mesothelin-related protein; MSGTs, malignant salivary gland tumors; MM, multiple myeloma; RRMM, recurrent and relapsed multiple myeloma.

 E-submission
E-submission