Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 58(3); 2024 > Article
-
Review
Interpretation of PD-L1 expression in gastric cancer: summary of a consensus meeting of Korean gastrointestinal pathologists -
Soomin Ahn1
 , Yoonjin Kwak2
, Yoonjin Kwak2 , Gui Young Kwon3
, Gui Young Kwon3 , Kyoung-Mee Kim1
, Kyoung-Mee Kim1 , Moonsik Kim4
, Moonsik Kim4 , Hyunki Kim5
, Hyunki Kim5 , Young Soo Park6
, Young Soo Park6 , Hyeon Jeong Oh7
, Hyeon Jeong Oh7 , Kyoungyul Lee8
, Kyoungyul Lee8 , Sung Hak Lee9
, Sung Hak Lee9 , Hye Seung Lee2
, Hye Seung Lee2
-
Journal of Pathology and Translational Medicine 2024;58(3):103-116.
DOI: https://doi.org/10.4132/jptm.2024.03.15
Published online: April 25, 2024
1Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
2Department of Pathology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
3Seoul Clinical Laboratories, Department of Pathology, Yongin, Korea
4Department of Pathology, School of Medicine, Kyungpook National University, Kyungpook National University Chilgok Hospital, Daegu, Korea
5Department of Pathology, Yonsei University College of Medicine, Seoul, Korea
6Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
7Department of Pathology, Seoul National University Bundang Hospital, Seongnam, Korea
8Pathology Center, Seegene Medical Foundation, Seoul, Korea
9Department of Hospital Pathology, Seoul St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- Corresponding Author: Hye Seung Lee, MD, PhD, Department of Pathology, Seoul National University Hospital, Seoul National University College of Medicine, 101 Daehak-ro, Jongno-gu, Seoul 03080, Korea Tel: +82-2-740-8269, Fax: +82-2-744-8273, E-mail: hye2@snu.ac.kr
© 2024 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
- Abstract
- CURRENT PRACTICE OF PD-L1 TESTING IN GASTRIC CANCER
- INTEROBSERVER CONCORDANCE OF PD-L1 COMBINED POSITIVE SCORE
- CONSENSUS MEETING OF KOREAN GASTROINTESTINAL PATHOLOGISTS
- CASES WITH CONSENSUS COMBINED POSITIVE SCORE <5
- CASES WITH CONSENSUS COMBINED POSITIVE SCORE ≥5
- CONCLUSION
- Supplementary Information
- NOTES
- REFERENCES
Abstract
- Nivolumab plus chemotherapy in the first-line setting has demonstrated clinical efficacy in patients with human epidermal growth factor receptor 2–negative advanced or metastatic gastric cancer, and is currently indicated as a standard treatment. Programmed death-ligand 1 (PD-L1) expression is an important biomarker for predicting response to anti–programmed death 1/PD-L1 agents in several solid tumors, including gastric cancer. In the CheckMate-649 trial, significant clinical improvements were observed in patients with PD-L1 combined positive score (CPS) ≥ 5, determined using the 28-8 pharmDx assay. Accordingly, an accurate interpretation of PD-L1 CPS, especially at a cutoff of 5, is important. The CPS method evaluates both immune and tumor cells and provides a comprehensive assessment of PD-L1 expression in the tumor microenvironment of gastric cancer. However, CPS evaluation has several limitations, one of which is poor interobserver concordance among pathologists. Despite these limitations, clinical indications relying on PD-L1 CPS are increasing. In response, Korean gastrointestinal pathologists held a consensus meeting for the interpretation of PD-L1 CPS in gastric cancer. Eleven pathologists reviewed 20 PD-L1 slides with a CPS cutoff close to 5, stained with the 28-8 pharmDx assay, and determined the consensus scores. The issues observed in discrepant cases were discussed. In this review, we present cases of gastric cancer with consensus PD-L1 CPS. In addition, we briefly touch upon current practices and clinical issues associated with assays used for the assessment of PD-L1 expression in gastric cancer.
- Table 1 summarizes the assays used for PD-L1 testing in GC. Currently, three standardized PD-L1 immunohistochemical assays (22C3 pharmDx, 28–8 pharmDx, and SP263) are used to specifically predict responses to pembrolizumab, nivolumab, and tislelizumab.
- In GC, PD-L1 expression is scored using the CPS scoring system, which is calculated as the number of PD-L1–stained cells (tumor cells, lymphocytes, and macrophages) divided by the total number of viable tumor cells multiplied by 100 [20]. A tumor cell is PD-L1 positive if the cell membrane is partially or completely stained, irrespective of staining intensity [20]. PD-L1–stained immune cells include only mononuclear inflammatory cells (lymphocytes or macrophages) within tumor nests and adjacent stroma and show membrane and/or cytoplasmic staining [20]. Other stromal cells such as fibroblasts, neutrophils, and plasma cells are excluded [20]. If the calculation result exceeds 100, it is presented as a maximum score of 100 [20]. If the PDL1 staining is heterogeneous, the final CPS is estimated by calculating the CPS results for each area within the entire tumor [20].
- Currently, two different PD-L1 assays are approved for clinical evaluation. For each assay, different CPS cutoff values are used. The PD-L1 22C3 pharmDx assay uses CPS ≥1 for PD-L1 positivity [8,10] and the 28-8 pharmDx assay uses CPS ≥5 [3]. For tislelizumab, tumor area positivity score, stained with SP263, is used [7]. Reports should specify the assay type and appropriate cutoff value used for the interpretation of PD-L1 positivity [15].
CURRENT PRACTICE OF PD-L1 TESTING IN GASTRIC CANCER
- Table 2 summarizes the studies that evaluated the interobserver concordance of PD-L1 CPS in GC. Two previous studies reported excellent interobserver agreement for PD-L1 evaluation in GC [13,21]. In a study by Kulangara et al. [13], three pathologists evaluated 68 22C3 pharmDx-stained PD-L1 slides (sample type not disclosed); an overall percentage agreement (OPA) of 96.6% at a CPS cutoff of 1 was observed. Nuti et al. [21] evaluated the interobserver concordance among 120 pathologists in their interpretation of 20 22C3 pharmDx-stained surgical samples; an OPA of 90.6% at the CPS cutoff of 1 was observed, with a Fleiss kappa value of 0.828.
- In contrast, Park et al. [22] reported poor interobserver concordance among five pathologists who evaluated 55 tissue microarray samples using the 22C3 pharmDx and SP263 assays. The intraclass correlation coefficient (ICC) was 0.387, and the Fleiss kappa value was 0.389 at a 22C3 pharmDx CPS cutoff of 1 [22]. In addition, two recent studies reported poor interobserver agreement regarding the CPS in GC [18,19]. Fernandez et al. [18] evaluated the concordance for PD-L1 CPS among 14 pathologists from 13 institutions in the United States, based on their assessment of 22C3 pharmDx-stained 112 biopsy samples. At a CPS cutoff of 1, the reported OPA and ICC were 31.48% and 0.484 [18], respectively. Higher CPS cutoffs (10 or 20) showed improved agreement than a CPS cutoff 1 [18]. Robert et al. [19] evaluated the interobserver agreement among 12 international pathologists who assessed 100 biopsy samples stained with PD-L1 28-8 and 22C3 pharmDx; interobserver CPS agreement was fair for both pre- (ICC range, 0.45 to 0.55) and post-training (ICC range, 0.56 to 0.57) for both assays [19]. In addition, Robert et al. [19] evaluated CPS interobserver agreement for 35 biopsy fragments. Poor or fair agreement was observed regarding the number of PD-L1–positive immune cells (ICC, 0.19), number of PD-L1–positive tumor cells (ICC, 0.54), total number of viable tumor cells (ICC, 0.09), and the calculated CPS (ICC, 0.14), whereas calculated tumor cell scores showed excellent agreement (ICC, 0.82) [19]. These findings are in agreement with the results for other solid tumors, wherein the interobserver concordance for immune cells was much lower than that for tumor cells [23-25].
INTEROBSERVER CONCORDANCE OF PD-L1 COMBINED POSITIVE SCORE
- Twenty GC samples stained using the PD-L1 28-8 pharmDx assay, in routine clinical pathology practice, with CPS scores ranging from 3 to 7, were collected from seven institutes in Korea; six were resection specimens and 14 were biopsy specimens. The information about samples is summarized in Table 3. PD-L1 slides were scanned using each institute’s scanner. Twelve specimens were scanned using a Philips Digital Pathology Slide SG300 scanner (Amsterdam, Netherlands), four with an Aperio GT 450 DX scanner (Vista, CA, USA), three with an Aperio AT2 scanner (Vista, CA, USA) and one with a 3DHISTECH PANNORAMIC 250 Flash III scanner (Budapest, Hungary). All the participating pathologists specialized in gastrointestinal pathology (the average number of years of experience, 15; range, 6 to 30 years) and routinely assessed PD-L1 28-8 pharmDx in GC in clinical practice. Nine work in university hospitals, and two work in private laboratories.
- On the day of the meeting (17 Oct 2023), 11 pathologists evaluated 20 PD-L1 scanned slides using each image viewer, without clinical information, for 30 min and submitted the CPS results. Some pathologists did not complete the survey for all the cases. Subsequently, all the pathologists reviewed the scanned PD-L1 image slides, discussed each case, and determined consensus CPS scores. For cases with poor interobserver agreement (more than 20% of pathologists disagreed with the CPS cutoff of 5), cases were discussed in detail, and a consensus CPS score was determined. Despite discussion, a CPS consensus was not reached for Case 3 (Supplementary Fig. S1).
CONSENSUS MEETING OF KOREAN GASTROINTESTINAL PATHOLOGISTS
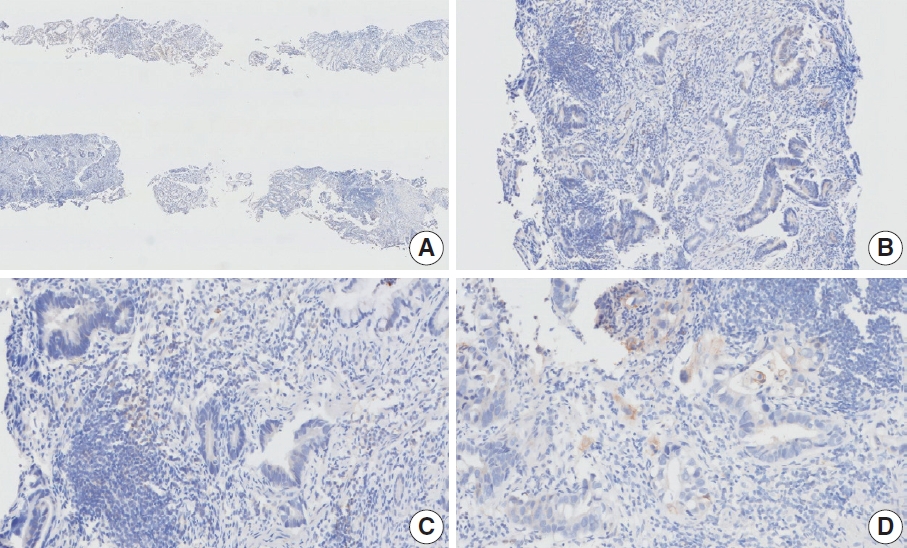
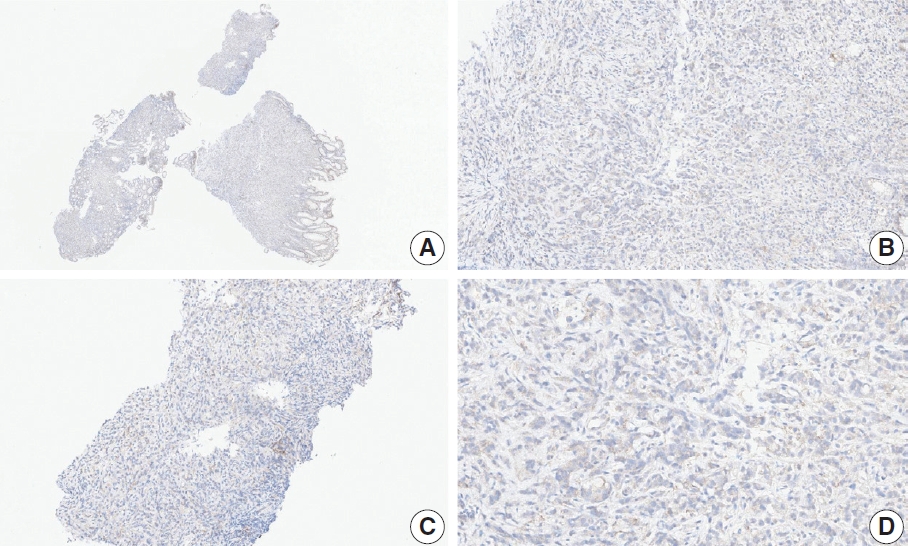
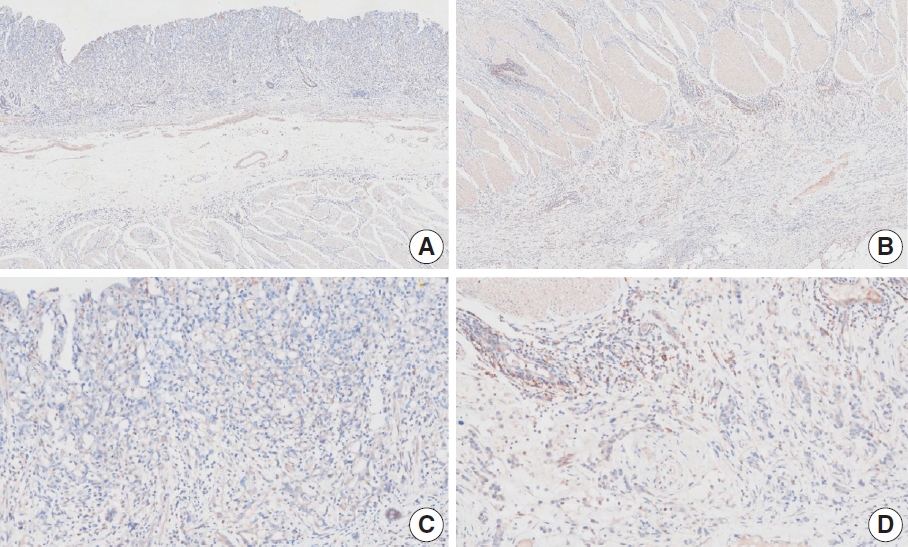
- Nine cases were concluded to have consensus CPS of <5. Of these, there was good interobserver agreement for two cases (cases 1 and 15) (Figs. 1, 2). The remaining cases are presented along with a detailed discussion.
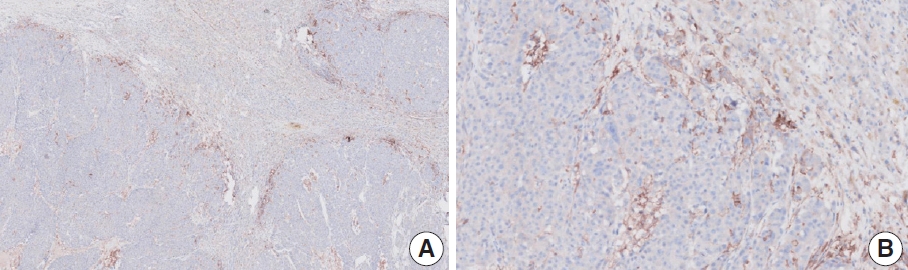
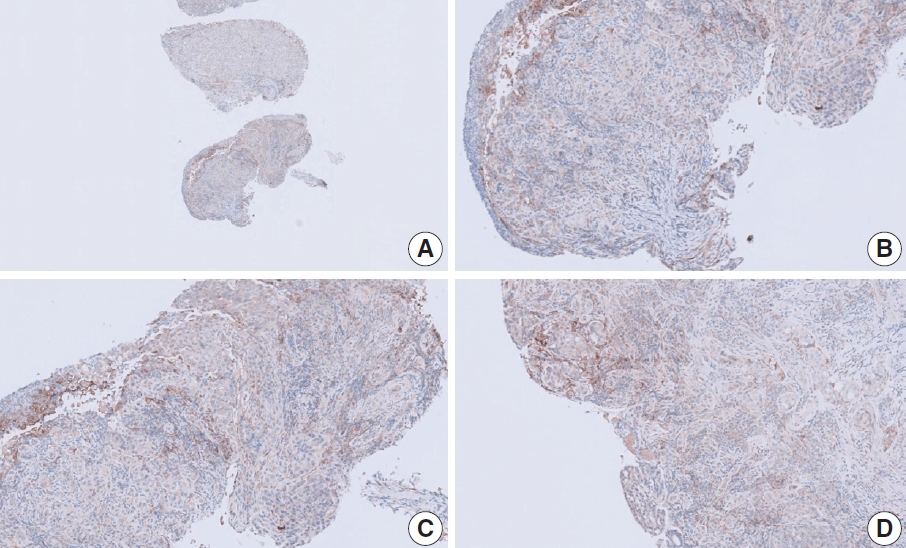
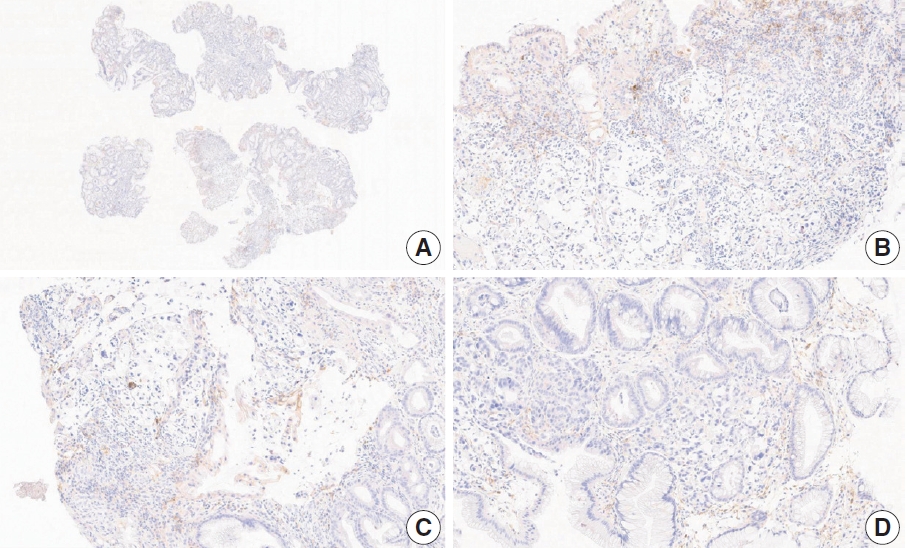
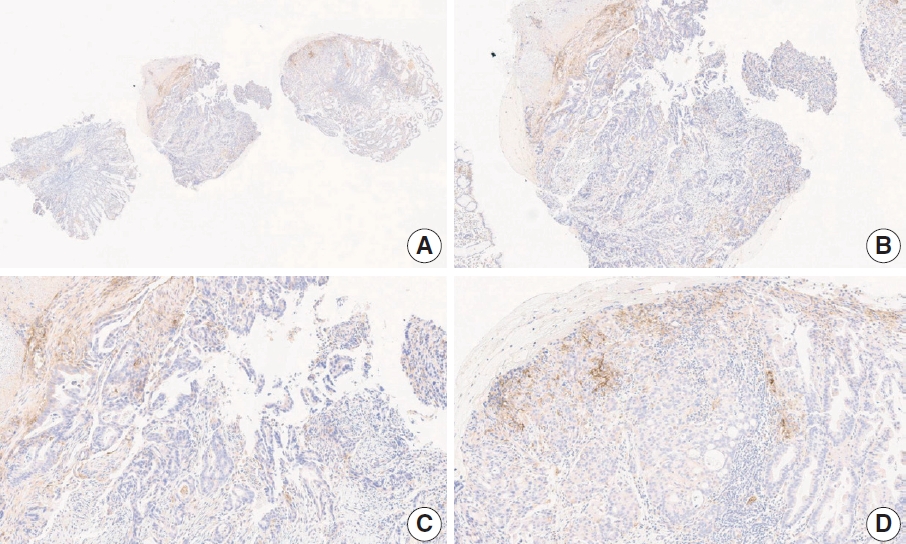
- Case 2 (Fig. 3)
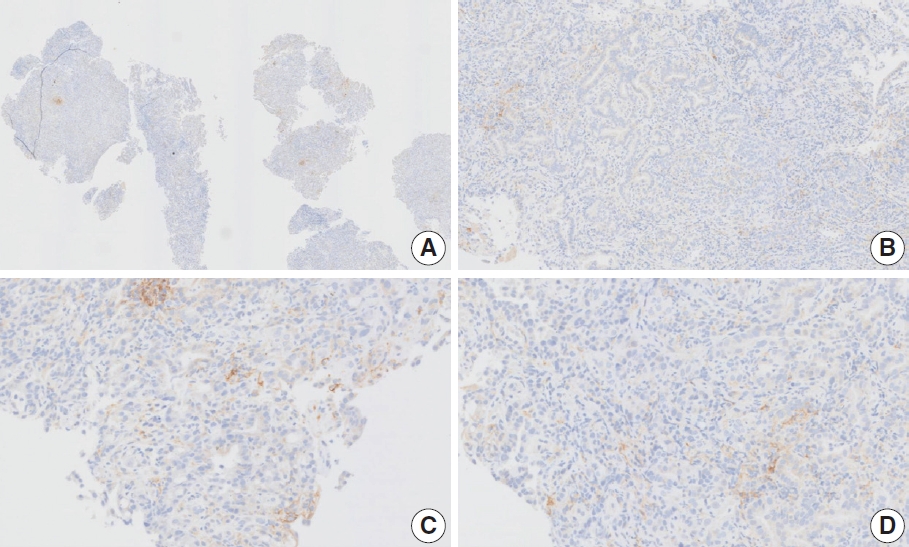
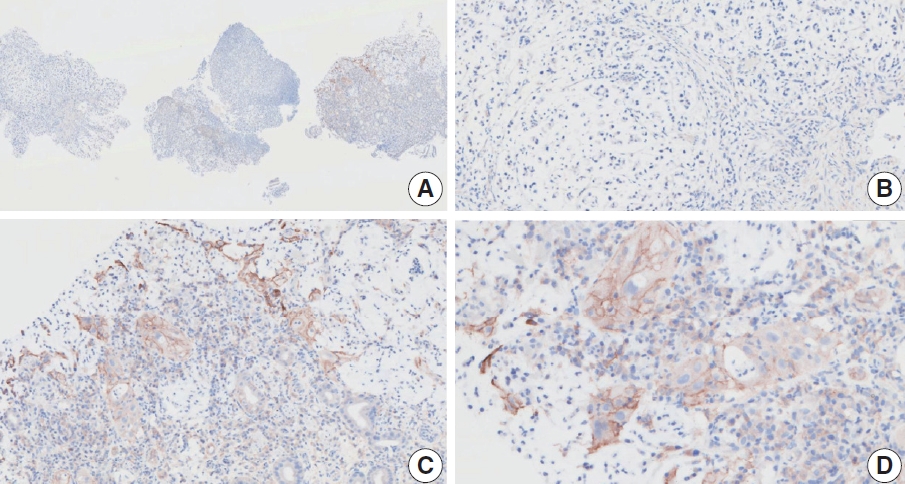
- At a CPS cutoff of 5, there was poor interobserver agreement (CPS 0, n=3; CPS 2, n=2; CPS 3, n=2; CPS 5, n=3). The biopsy specimen consisted of three pieces of signet ring cell carcinoma with normal components. One piece was PD-L1 negative, the second exhibited stained area scores less than 5, and the third one exhibited stained area scores of >5. When the PD-L1 staining revealed a heterogeneous pattern, the final CPS was estimated for each area [20]. By averaging the scores for each region, a consensus CPS score of 2 was determined.
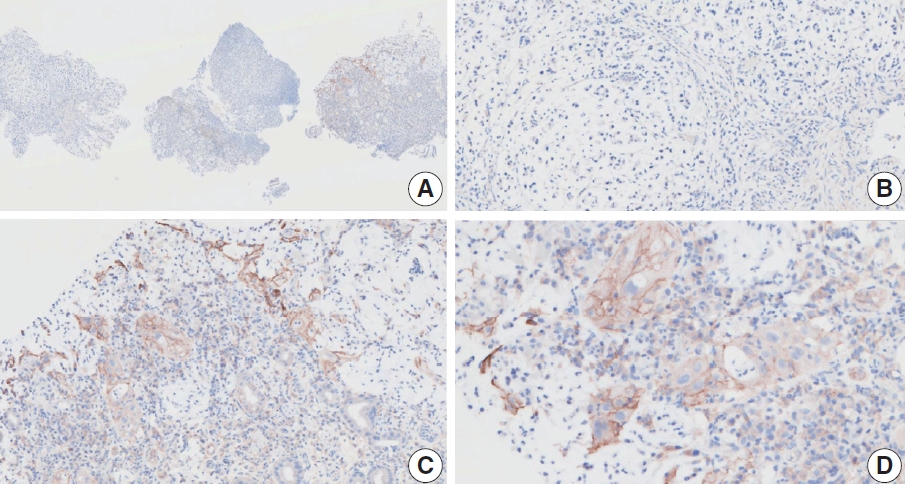
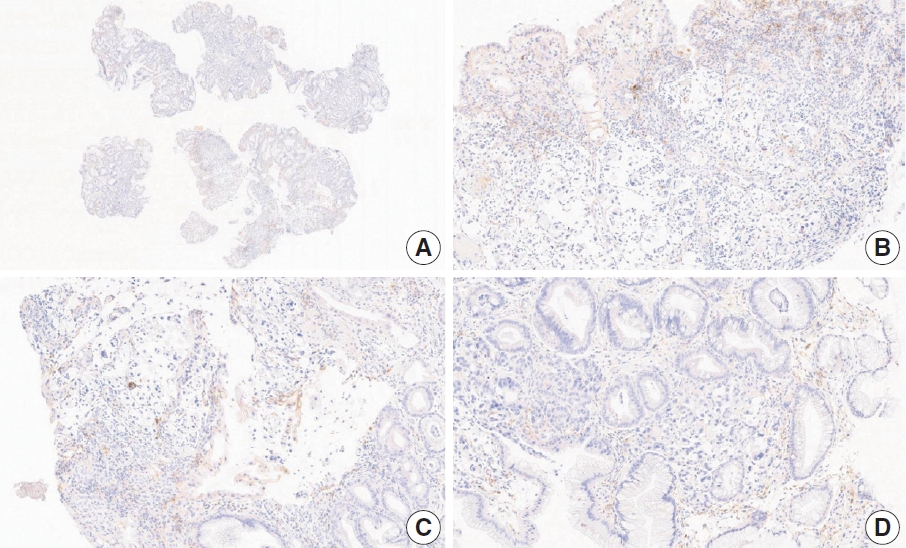
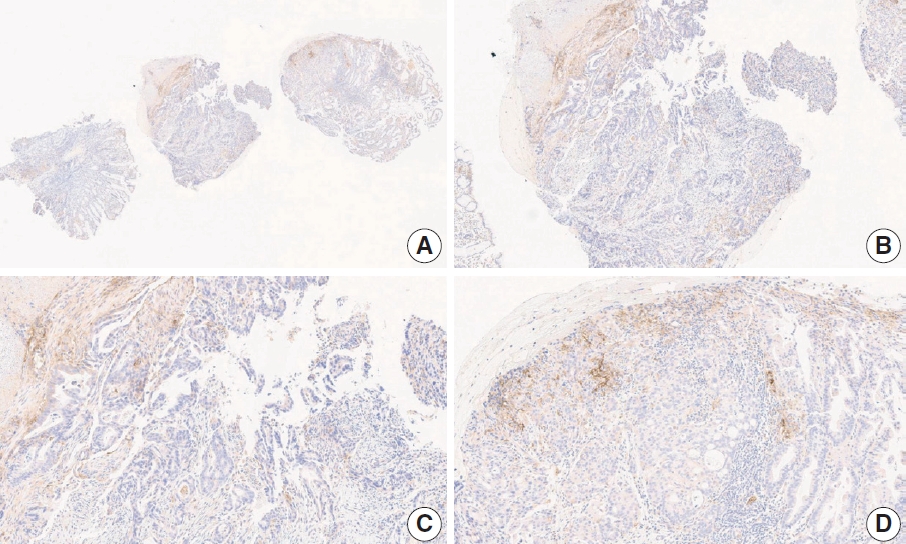
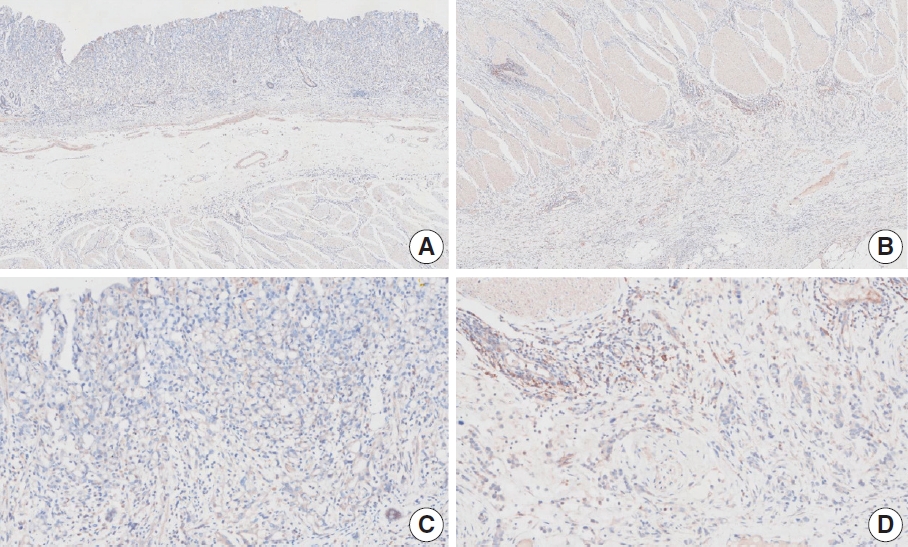
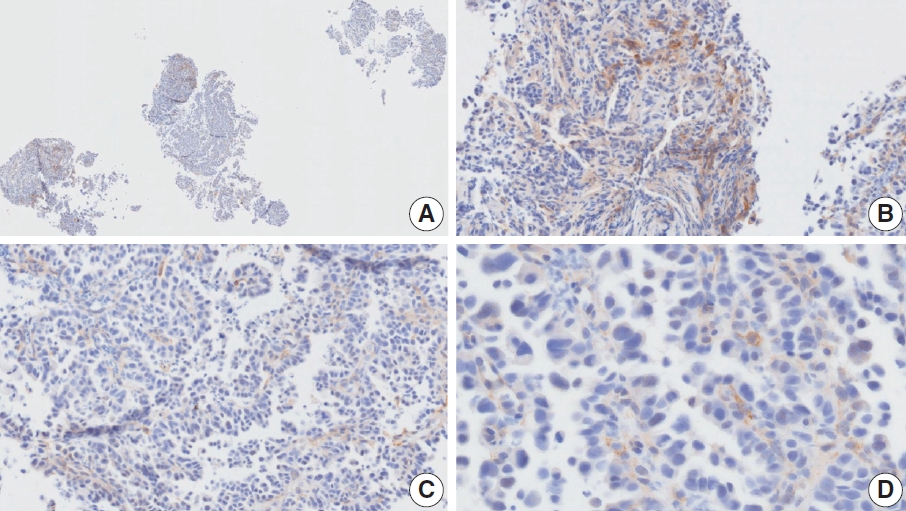
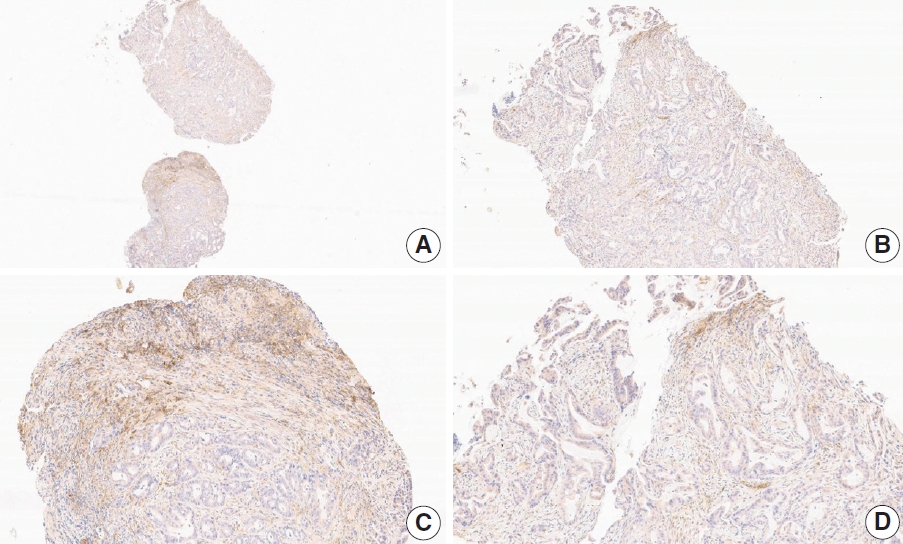
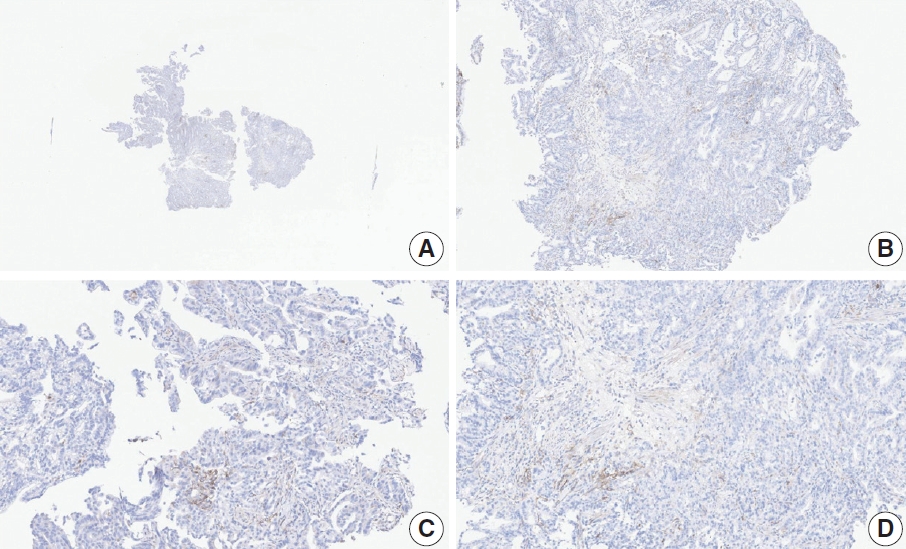
- Case 6 (Fig. 4)
- At a CPS cutoff of 5, there was poor interobserver agreement (CPS 1, n=1; CPS 2, n=2; CPS 3, n=1; CPS 5, n=4; CPS 8, n=1; CPS 10, n=1). This biopsy specimen consisted of two pieces of moderately differentiated adenocarcinoma tissue without a normal component. One fragment had no PD-L1–positive cells, while the other showed stained area scores of CPS >5. A consensus CPS score of 3 was determined by averaging the scores for each region.
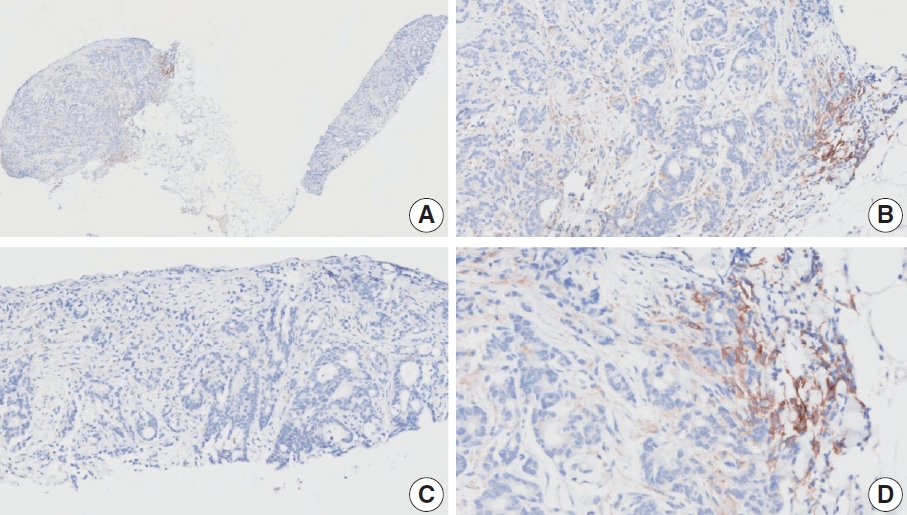
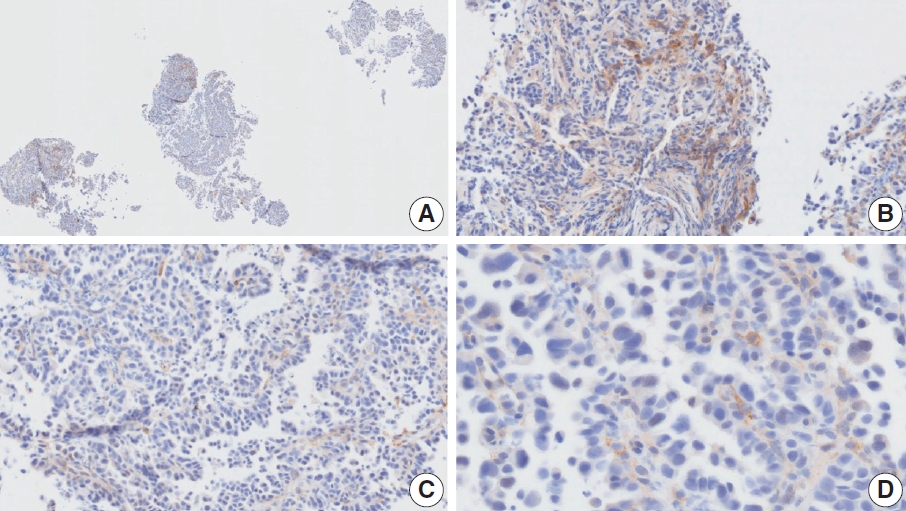
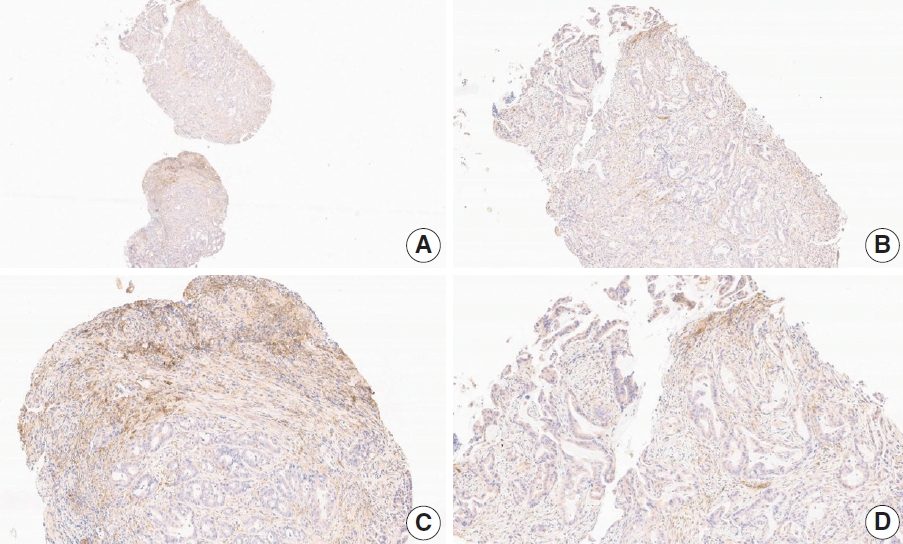
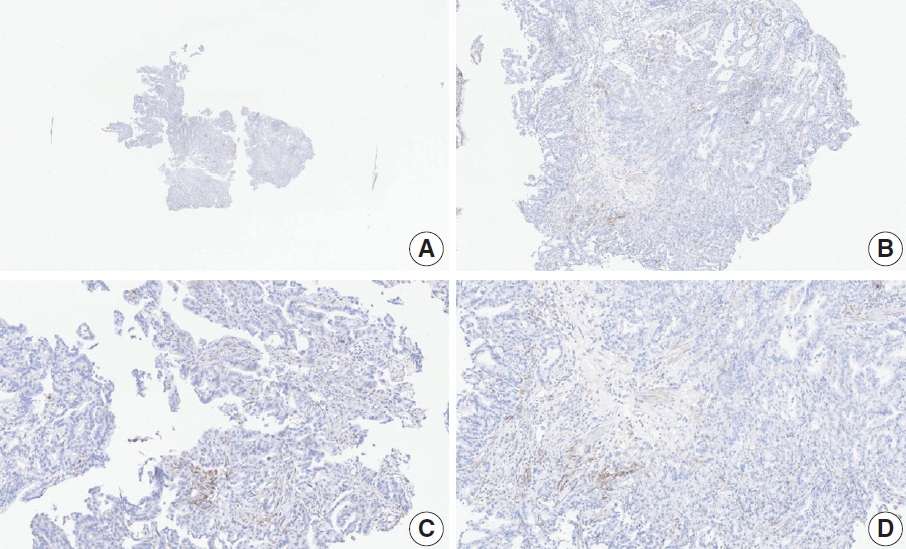
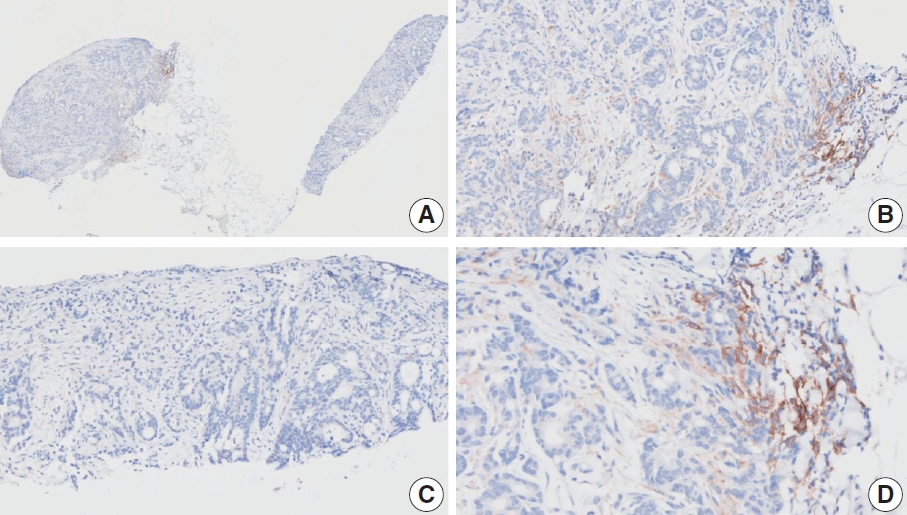
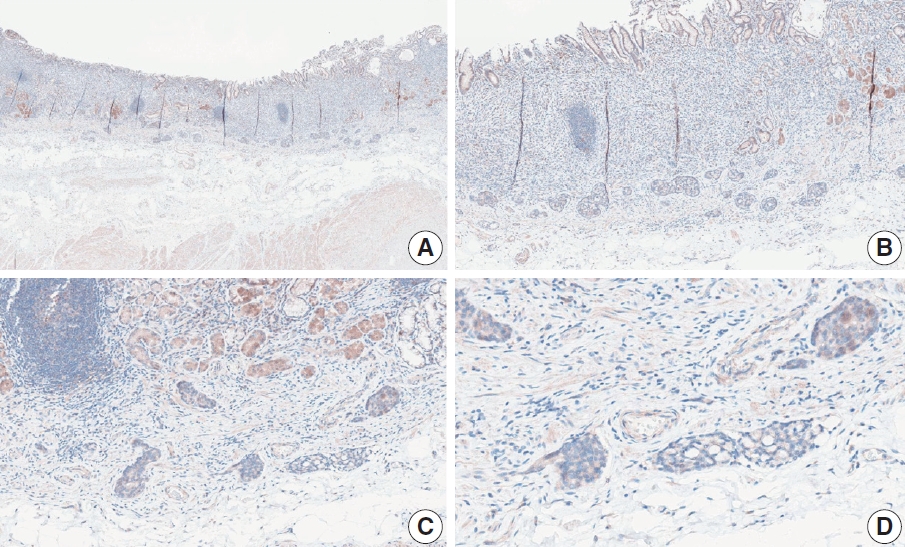
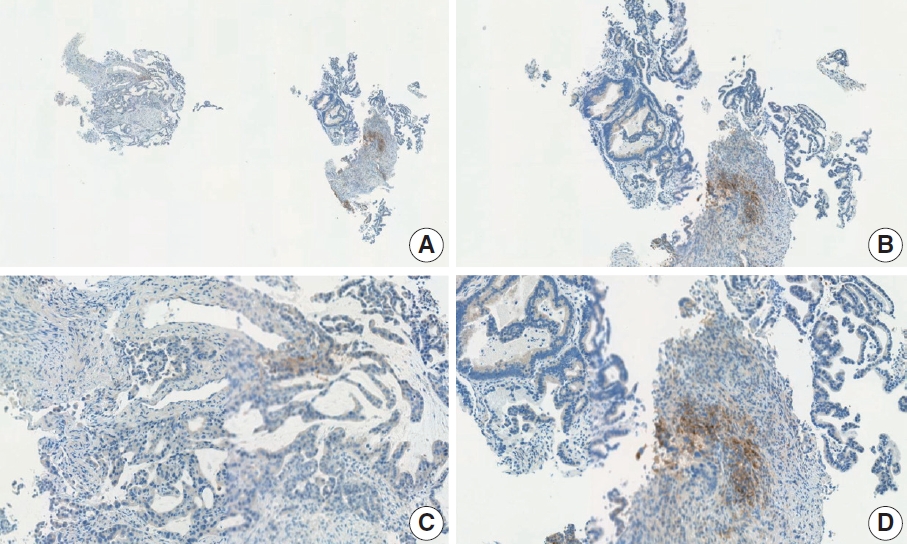
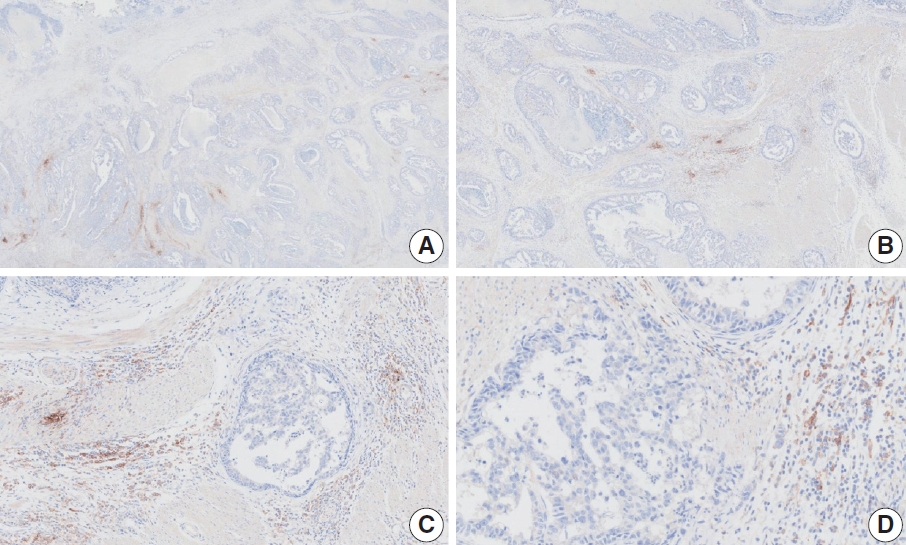
- Case 7 (Fig. 5)
- At a CPS cutoff of 5, there was poor interobserver agreement (CPS 0, n=2; CPS 3, n=2; CPS 4, n=1; CPS 5, n=1; CPS 7, n=1; CPS 9, n=1). The biopsy specimen comprised four fragments of poorly cohesive carcinoma with normal components. While most pieces were PD-L1–negative, one piece revealed PD-L1–positive immune cells. On an average, a consensus score of 2 was determined.
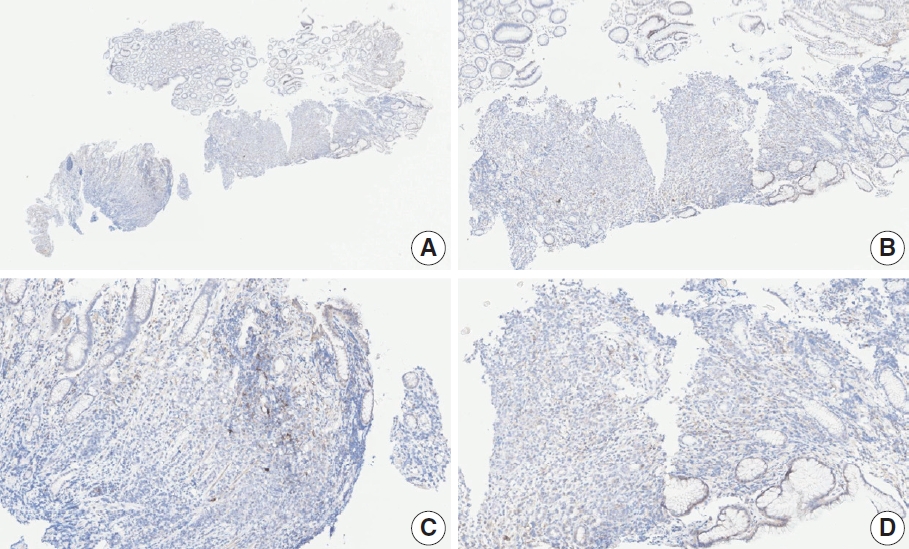
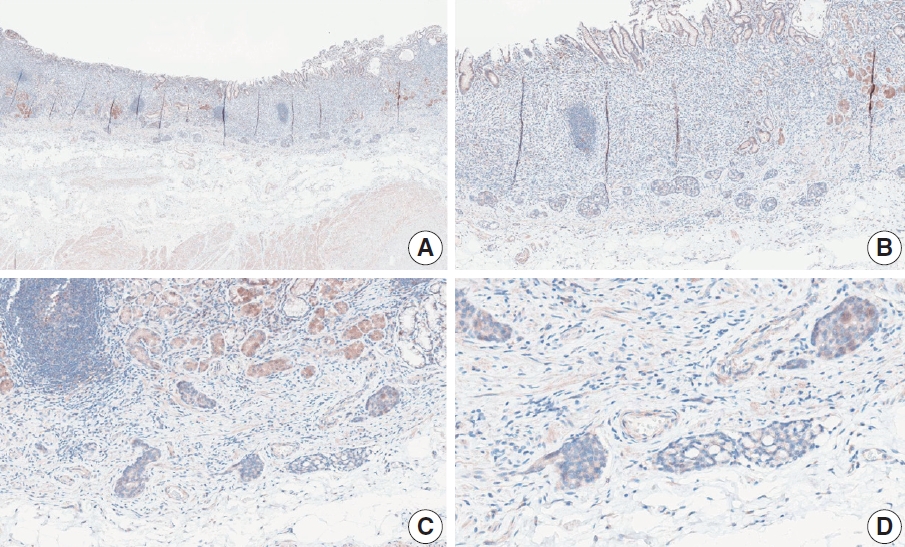
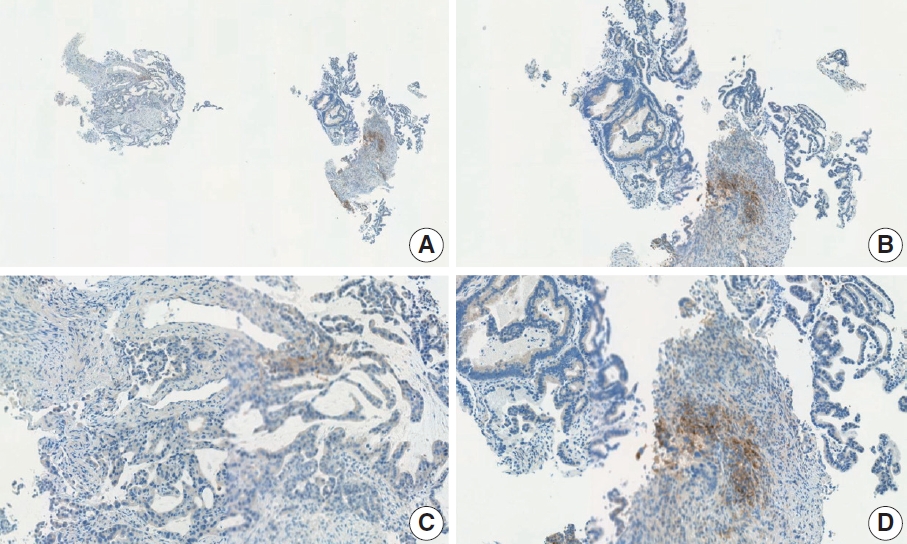
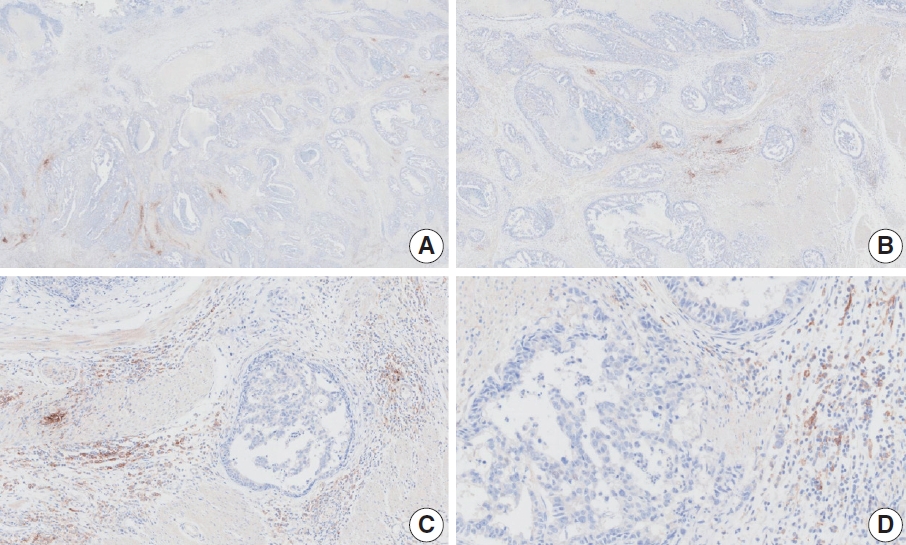
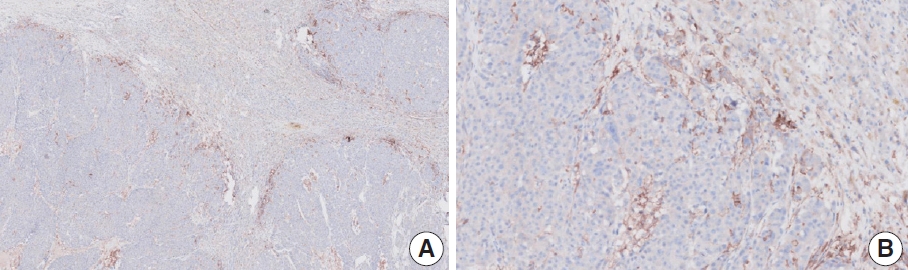
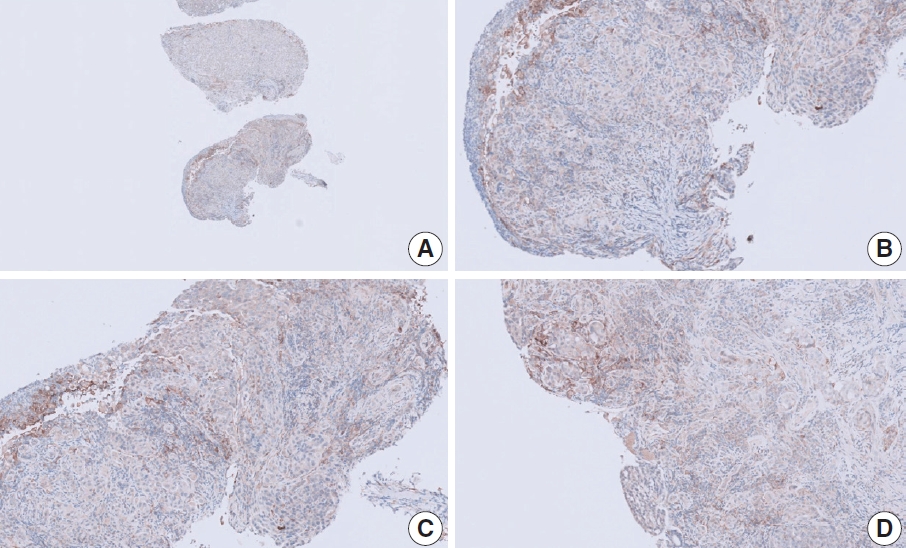
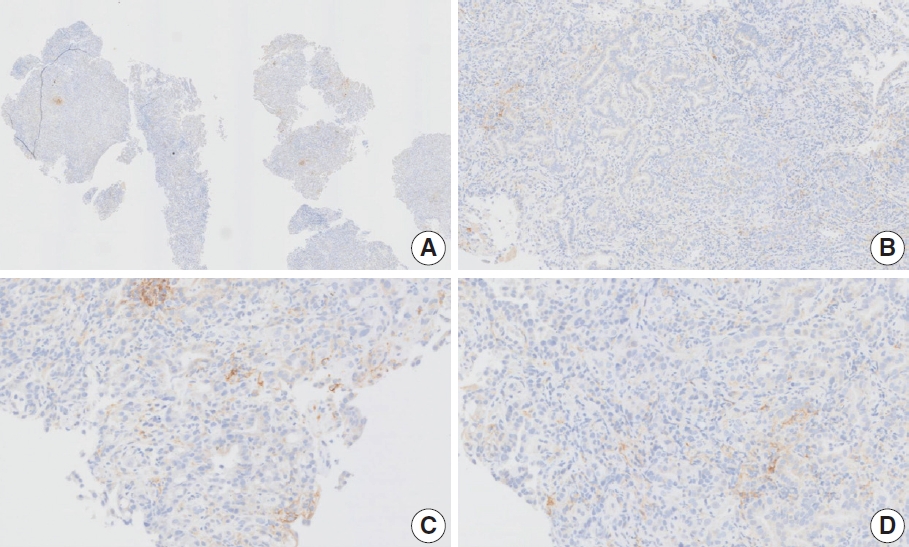
- Case 9 (Fig. 6)
- The biopsy specimen contained a poorly differentiated adenocarcinoma with infiltrating immune cells within and adjacent to the tumor. Most pathologists submitted CPS results of <5 (CPS 0, n=1; CPS 1, n=2; CPS 2, n=1; CPS 3, n=1; CPS 5, n=2); following a detailed discussion, a CPS consensus of 2 was reached. Many pathologists at the meeting mentioned that nonspecific 28-8 pharmDx staining complicates CPS interpretation. Although the tumor cells showed weak staining at a low magnification, only cytoplasmic staining was observed at a high magnification. Tumor cells with cytoplasmic staining only need to be excluded from CPS calculations [20]. In this case, only PD-L1–positive lymphocytes were included in the CPS numerator. Nonspecific cytoplasmic staining in tumor cells is not usually observed in the 22C3 pharmDx assay, and caution is required for interpretation of 28-8 pharmDx assay [17].
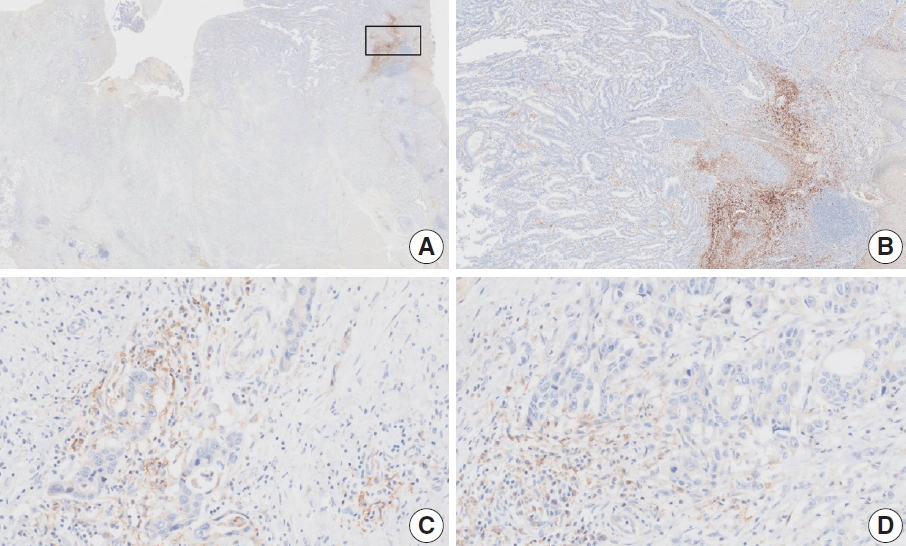
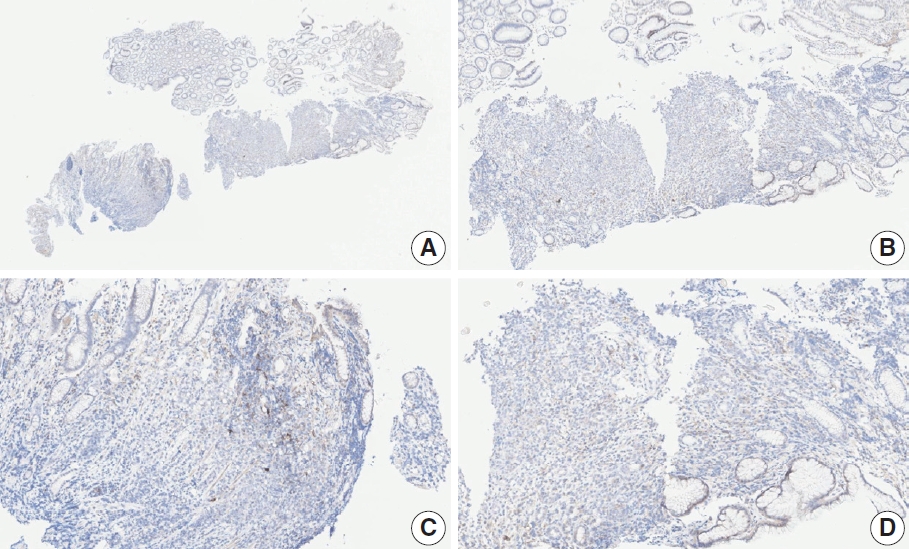
- Case 11 (Fig. 7)
- This case revealed poor interobserver agreement at a CPS cutoff of 5 (CPS 0, n=1; CPS 3, n=2; CPS, n=4). Six of seven fragments of the biopsy specimen contained signet ring cell carcinoma with the extracellular mucin. Some tumor cells showed partial membrane staining, and lymphocytes showed cytoplasmic staining. Nonspecific staining in the empty space caused by mucin, stained stromal cells, normal glandular cells stained by edge artifacts, and neutrophils were excluded from the CPS calculation. In this case, defining the tumor-associated area was a confounding factor for interpretation, and it was difficult to determine whether the immune cells were within the tumor or distant from the tumor. In general, a 20× field of view rule is applied to define tumor-associated areas [20]. Immune cells are considered tumor-associated and included in the CPS calculation if the cells are present within the tumor nests or adjacent stroma or both within a 20× magnification field of view [20]. At the same time, non-tumor gastritis areas need to be excluded [20]. However, endoscopic specimens inevitably contain non-tumor gastritis areas wherein resident inflammation is common [19]. In this case, when the 20× field of view rule was applied, the surface area presumed to be non-tumor gastritis was included. This confused the pathologists, as there are currently no guidelines for this situation. Considering that mucinous tumors are generally less immunogenic, and immune cells in the specimen are considered mostly gastritis, pathologists at the meeting agreed to a CPS of 3. A correlation with the treatment response would be helpful; however, the patient had not yet received immunotherapy.
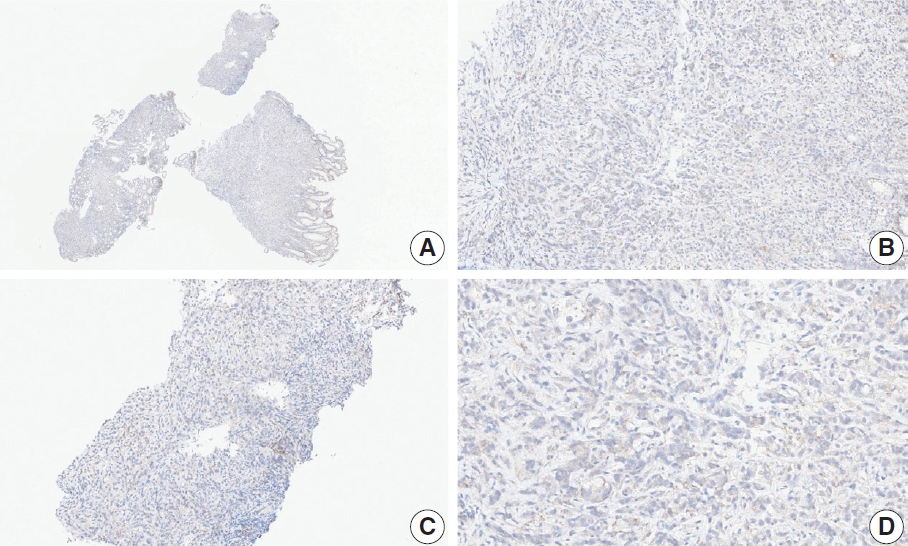
- Case 17 (Fig. 8)
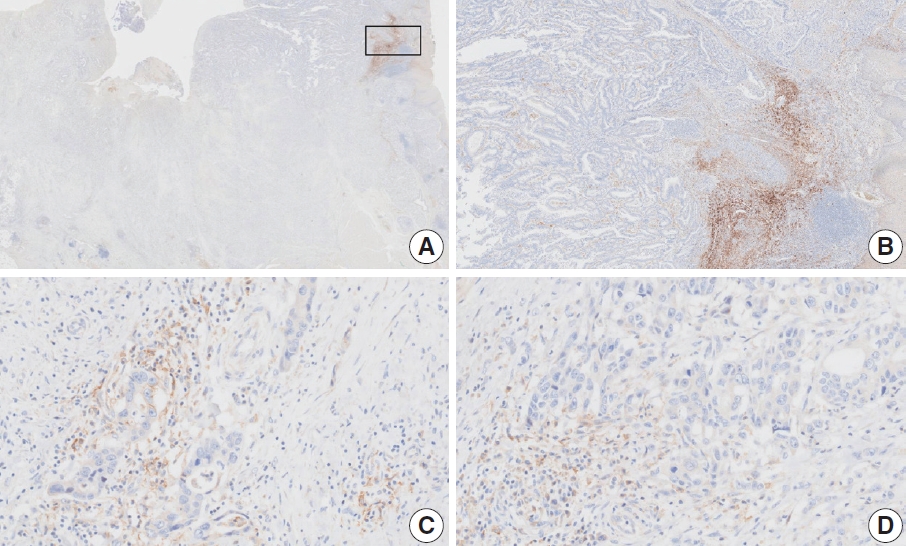
- At a CPS cutoff of 5, there was poor interobserver agreement (CPS 2, n=1; CPS 3, n=1; CPS 4, n=1; CPS 5, n=2; CPS 10, n=1; CPS 20, n=1). The tumor histology was a papillary adenocarcinoma with empty spaces caused by the papillary architecture. At a low magnification, some stained areas were observed. However, at a high magnification, most of the stained areas were revealed as background staining in empty spaces and stromal cells. After excluding nonspecific background staining, the number of positive immune cells appeared to be small. After discussion, the pathologists agreed on a CPS of 2.
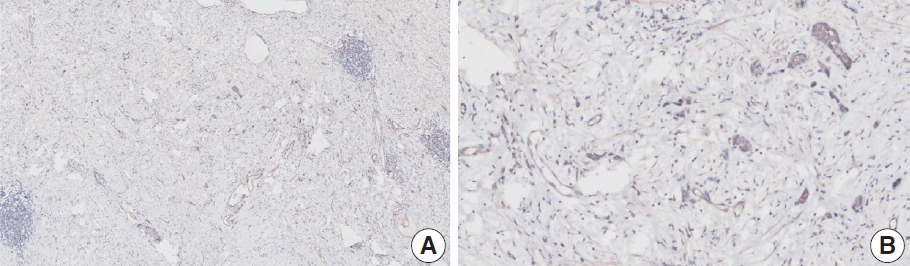
- Case 20 (Fig. 9)
- The resected specimen showed a poorly cohesive carcinoma with abundant lymphatic tumor emboli in the mucosa and submucosa. The submitted results were CPS 0 (n=1) and CPS 5 (n=2). Tumor cells show only cytoplasmic staining, which must be excluded from the CPS calculation [20]. Normal glandular cells exhibited nonspecific staining. The pathologists agreed on a CPS of 0.
CASES WITH CONSENSUS COMBINED POSITIVE SCORE <5
- Ten cases were concluded as having consensus CPS ≥5. Among these, good interobserver agreement was noted for five cases (cases 5, 10, 12, 18, and 19) (Figs. 10, 11, 12, 13, and 14, respectively). The remaining cases are presented along with a detailed discussion.
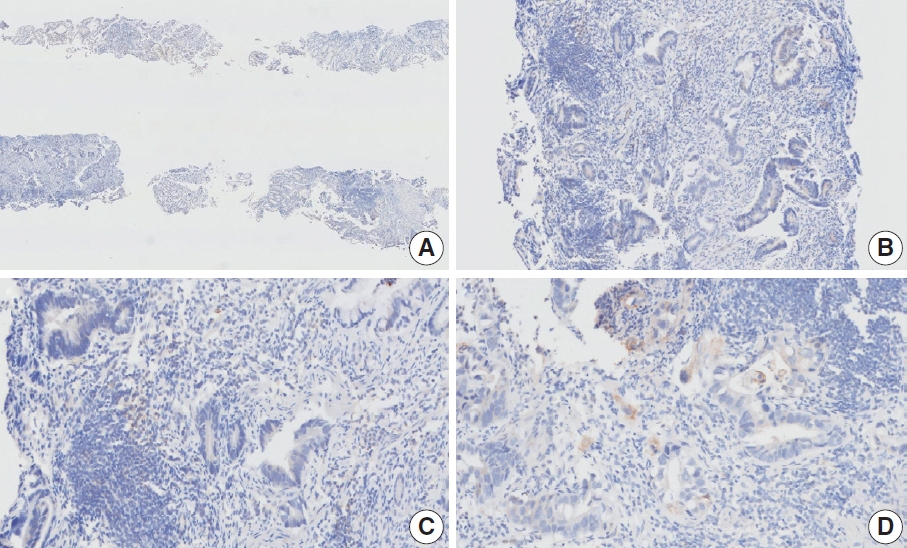
- Case 4 (Fig. 15)
- At a CPS cutoff of 5, there was poor interobserver agreement (CPS 0, n=3; CPS 1, n=1; CPS 2, n=1; CPS 5, n=2; CPS 8, n =1; CPS 10, n=1; CPS 20, n=1). This resected specimen consisted of poorly cohesive carcinoma with some infiltrating lymphocytes. Owing to the large tumor area, CPS scoring was difficult. Compared to those in the usual diffuse-type GC, relatively abundant tumor-associated lymphocytes were observed in the specimen, and these immune cells were focally PD-L1 positive. Based on the findings, a consensus CPS of 5 was determined.
- Case 8 (Fig. 16)
- At a CPS cutoff of 5, there was poor interobserver agreement (CPS 1, n=1; CPS 2, n=2; CPS 3, n=1; CPS 5, n=1; CPS 7, n=1). This tumor consisted of four papillary adenocarcinoma fragments and two normal mucosal fragments. Nonspecific staining was observed in the empty space and immune cells in the non-tumor gastritis area. However, even after the exclusion of nonspecific staining, PD-L1–positive immune cells remained around the tumor. A consensus CPS of 5 was determined.
- Case 13 (Fig. 17)
- At a CPS cutoff of 5, there was poor interobserver agreement (CPS 2, n=1; CPS 3, n=2; CPS 4, n=2; CPS 5, n=3; CPS 6, n=1; CPS 10, n=1). This resection specimen of advanced GC exhibited a small PD-L1–stained area at the front of the invasive tumor. Although the PD-L1–positive area was small, some of the pathologists mentioned that there were abundant immune cells, and PD-L1–positive immune cells at the invasive front were likely the result of a direct interaction between tumor cell antigens and neighboring immune cells. The final consensus was a CPS of 5. The interpretation manual provides guidelines for the calculation of CPS in a small PD-L1–stained area [20]. First, the percentage of the PD-L1–stained area in the entire tumor was evaluated [20]. Next, the CPS of the area was evaluated via staining [20]. Finally, the CPS was calculated considering both the results [20]. By applying this rule, 10% of the entire area was deemed PD-L1 positive, and the CPS was 50. The CPS of the entire tumor area was 10%×50=CPS 5.
- Case 14 (Fig. 18)
- At a CPS cutoff of 5, there was poor interobserver agreement (CPS 1, n=2; CPS 2, n=1; CPS 3, n=1; CPS 5, n=2; CPS 8, n=1; CPS 10, n=1). The biopsy specimen consisted of tumor cells and abundant tumor-infiltrating lymphocytes without a normal component. A few tumor and immune cells with strong intensity were observed in a small area. Furthermore, a considerable number of weakly stained cells was observed. The staining intensity can vary significantly in a single case [26]. Distinguishing positive staining with weak intensity from nonspecific staining was challenging. Despite background staining, immune cells with membrane and/or cytoplasmic staining at any intensity should be included in CPS calculation. Therefore, by counting the immune cells (not all but some) with cytoplasmic staining, this case was classified as CPS 5.
- Case 16 (Fig. 19)
- At a CPS cutoff of 5, there was poor interobserver agreement (CPS 2, n=1; CPS 3, n=1; CPS 5, n=2; CPS 6, n=1; CPS 7, n=1; CPS 8, n=1). Strongly stained areas were observed on mucosal surfaces. The ulcerative area of the mucosa was excluded from CPS calculation. However, the stained area (Fig. 19A box, B) in the mucosa contained tumors admixed with inflammatory cells and focal necrosis, which needed to be counted. In addition, positive intra-glandular macrophages, lymphocytes, and tumor cells were observed at the invasive front of the tumor. If PD-L1 staining shows heterogeneous results, the final CPS must be estimated by calculating the CPS result for each area [20]. Finally, a CPS of 5 was determined for this case.
CASES WITH CONSENSUS COMBINED POSITIVE SCORE ≥5
- Poor interobserver agreement among pathologists’ CPS in PD-L1 assessment is a known challenge for the use of PD-L1 as a biomarker in GC. In particular, CPS scoring in biopsy specimens stained with 28-8 pharmDx can be more challenging due to nonspecific staining and various artifacts. To address this, Korean gastrointestinal pathologists had a consensus meeting of PD-L1 in GC to discuss cases near CPS cutoff 5. Our consensus meeting also revealed high variability even among expert pathologists in some cases. In this review, we described cases with consensus CPS including practical considerations and potential pitfalls, which may serve as reference for pathologists.
CONCLUSION
Supplementary Information
Ethics Statement
Not applicable.
Availability of Data and Material
The datasets generated or analyzed during the study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
Author Contributions
Conceptualization: SA, HSL, KMK. Participation of consensus meeting: all authors. Supervision: HSL, KMK. Resources: YSP, YK, SA, KL, GYK, MK, HJO. Writing—original draft preparation: SA, HSL. Writing—review & editing: all authors. Approval of final manuscript: all authors.
Conflicts of Interest
S.H.L., a contributing editor of the Journal of Pathology and Translational Medicine, was not involved in the editorial evaluation or decision to publish this article. All remaining authors have declared no conflicts of interest.
Funding Statement
No funding to declare.
| PD-L1 assay | Cutoff | No. of observers | No. of cases | Sample type | Interobserver agreement | Fleiss kappa value | Reference |
|---|---|---|---|---|---|---|---|
| 22C3 PharmDx | CPS ≥ 1 | 3 | 68 | Not mentioned | OPA 96.6% | - | [13] |
| 22C3 PharmDx | CPS ≥ 1 | 120 | 20 (day1), 25 (day2) | Resection | OPA 90.6% | 0.828 | [21] |
| 22C3 PharmDx | CPS value | 5 | 55 | Tissue microarray | ICC 0.387 (lower 95% CI, 20.9%) | CPS ≥ 1 0.389 | [22] |
| SP263 | CPS value | 5 | 55 | Tissue microarray | ICC 0.349 (lower 95% CI, 13.5%) | CPS ≥ 1 0.256 | |
| 22C3 PharmDx | CPS ≥ 1 | 14 | 112 | Biopsy | OPA 31.48% (95% CI, 22.72–40.24) | 0.477 | [18] |
| ICC 0.484 (95% CI, 0.403–0.571) | - | ||||||
| CPS ≥ 10 | 14 | 112 | Biopsy | OPA 67.59% (95% CI, 58.77–76.42) | 0.607 | ||
| ICC 0.604 (95% CI, 0.584–0.624) | - | ||||||
| CPS ≥ 20 | 14 | 112 | Biopsy | OPA 83.33% (95% CI, 76.3–90.36) | 0.626 | ||
| ICC 0.629 (95% CI, 0.562–0.698) | - | ||||||
| 28-8 PharmDx | CPS value | 12 | 100 | Biopsy | ICC 0.45 (95% CI, 0.38–0.53) | - | [19] |
| 22C3 PharmDx | CPS value | 12 | 100 | Biopsy | ICC 0.55 (95% CI, 0.47–0.63) | - |
| Case No. | Institution | Vendor/file format | Specimen type | Interobserver agreement | Consensus CPS |
|---|---|---|---|---|---|
| 1 | A | Philips/.isyntax | Resection | Gooda | 0 |
| 2 | A | Philips/.isyntax | Biopsy | Poor | 2 |
| 3 | A | Philips/.isyntax | Biopsy | Poor | Not determined |
| 4 | A | Philips/.isyntax | Resection | Poor | 5 |
| 5 | A | Philips/.isyntax | Resection | Good | 5 |
| 6 | A | Philips/.isyntax | Biopsy | Poor | 3 |
| 7 | B | Leica Biosystems/.svs | Biopsy | Poor | 2 |
| 8 | B | Leica Biosystems/.svs | Biopsy | Poor | 5 |
| 9 | B | Leica Biosystems/.svs | Biopsy | Poor | 2 |
| 10 | C | Leica Biosystems/.svs | Biopsy | Good | ≥ 5 |
| 11 | C | Leica Biosystems/.svs | Biopsy | Poor | 3 |
| 12 | C | Leica Biosystems/.svs | Biopsy | Good | ≥ 5 |
| 13 | D | Philips/.isyntax | Resection | Poor | 5 |
| 14 | D | Philips/.isyntax | Biopsy | Poor | 5 |
| 15 | D | Philips/.isyntax | Biopsy | Good | 2 |
| 16 | D | Philips/.isyntax | Resection | Poor | 5 |
| 17 | D | Philips/.isyntax | Biopsy | Poor | 2 |
| 18 | E | 3DHistech/.MRXS | Biopsy | Good | 5 |
| 19 | F | Philips/.isyntax | Biopsy | Good | ≥ 5 |
| 20 | G | Leica Biosystems/.svs | Resection | Poor | 0 |
- 1. Ribas A. Tumor immunotherapy directed at PD-1. N Engl J Med 2012; 366: 2517-9. ArticlePubMed
- 2. Nakamura Y, Kawazoe A, Lordick F, Janjigian YY, Shitara K. Biomarker-targeted therapies for advanced-stage gastric and gastrooesophageal junction cancers: an emerging paradigm. Nat Rev Clin Oncol 2021; 18: 473-87. ArticlePubMedPDF
- 3. Janjigian YY, Shitara K, Moehler M, et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): a randomised, open-label, phase 3 trial. Lancet 2021; 398: 27-40. ArticlePubMedPMC
- 4. Kang YK, Chen LT, Ryu MH, et al. Nivolumab plus chemotherapy versus placebo plus chemotherapy in patients with HER2-negative, untreated, unresectable advanced or recurrent gastric or gastro-oesophageal junction cancer (ATTRACTION-4): a randomised, multicentre, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2022; 23: 234-47. ArticlePubMed
- 5. Rha SY, Oh DY, Yanez P, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for HER2-negative advanced gastric cancer (KEYNOTE-859): a multicentre, randomised, doubleblind, phase 3 trial. Lancet Oncol 2023; 24: 1181-95. PubMed
- 6. Shitara K, Van Cutsem E, Bang YJ, et al. Efficacy and safety of pembrolizumab or pembrolizumab plus chemotherapy vs chemotherapy alone for patients with first-line, advanced gastric cancer: the KEYNOTE-062 phase 3 randomized clinical trial. JAMA Oncol 2020; 6: 1571-80. ArticlePubMed
- 7. Xu J, Kato K, Raymond E, et al. Tislelizumab plus chemotherapy versus placebo plus chemotherapy as first-line treatment for advanced or metastatic oesophageal squamous cell carcinoma (RATIONALE-306): a global, randomised, placebo-controlled, phase 3 study. Lancet Oncol 2023; 24: 483-95. ArticlePubMed
- 8. Janjigian YY, Kawazoe A, Bai Y, et al. Pembrolizumab plus trastuzumab and chemotherapy for HER2-positive gastric or gastro-oesophageal junction adenocarcinoma: interim analyses from the phase 3 KEYNOTE-811 randomised placebo-controlled trial. Lancet 2023; 402: 2197-208. ArticlePubMed
- 9. Shitara K, Ajani JA, Moehler M, et al. Nivolumab plus chemotherapy or ipilimumab in gastro-oesophageal cancer. Nature 2022; 603: 942-8. ArticlePubMedPMCPDF
- 10. Fuchs CS, Doi T, Jang RW, et al. Safety and efficacy of pembrolizumab monotherapy in patients with previously treated advanced gastric and gastroesophageal junction cancer: phase 2 clinical KEYNOTE-059 trial. JAMA Oncol 2018; 4: e180013. ArticlePubMedPMC
- 11. Kubota Y, Aoki Y, Kawazoe A, Shitara K. Role of nivolumab in the management of first-Line unresectable advanced or recurrent gastric cancer in combination with chemotherapy: lessons from the Japanese experience. Cancer Manag Res 2022; 14: 3083-94. ArticlePubMedPMCPDF
- 12. Yi M, Jiao D, Xu H, et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer 2018; 17: 129.ArticlePubMedPMCPDF
- 13. Kulangara K, Zhang N, Corigliano E, et al. Clinical utility of the combined positive score for programmed death ligand-1 expression and the approval of pembrolizumab for treatment of gastric cancer. Arch Pathol Lab Med 2019; 143: 330-7. ArticlePubMedPDF
- 14. Lei M, Siemers NO, Pandya D, et al. Analyses of PD-L1 and inflammatory gene expression association with efficacy of nivolumab +/- ipilimumab in gastric cancer/gastroesophageal junction cancer. Clin Cancer Res 2021; 27: 3926-35. ArticlePubMedPDF
- 15. Park YS, Kook MC, Kim BH, et al. A standardized pathology report for gastric cancer: 2nd edition. J Pathol Transl Med 2023; 57: 1-27. ArticlePubMedPMCPDF
- 16. Kim TH, Kim IH, Kang SJ, et al. Korean Practice Guidelines for Gastric Cancer 2022: an evidence-based, multidisciplinary approach. J Gastric Cancer 2023; 23: 3-106. PubMedPMC
- 17. Ahn S, Kim KM. PD-L1 expression in gastric cancer: interchangeability of 22C3 and 28-8 pharmDx assays for responses to immunotherapy. Mod Pathol 2021; 34: 1719-27. ArticlePubMedPDF
- 18. Fernandez AI, Robbins CJ, Gaule P, et al. Multi-institutional study of pathologist reading of the programmed cell death ligand-1 combined positive score immunohistochemistry assay for gastric or gastroesophageal junction cancer. Mod Pathol 2023; 36: 100128.ArticlePubMedPMC
- 19. Robert ME, Ruschoff J, Jasani B, et al. High interobserver variability among pathologists using combined positive score to evaluate PDL1 expression in gastric, gastroesophageal junction, and esophageal adenocarcinoma. Mod Pathol 2023; 36: 100154.ArticlePubMed
- 20. PD-L1 IHC 28-8 pharmDx interpretation manual: gastric adenocarcinoma, gastroesophageal juction (GEJ) adenocarcinoma, and esophageal adenocarcinoma [Internet]. Santa Clara: DAKO Agilent Technologies, 2021 [cited 2024 Feb 22]. Available from: https://www.agilent.com/cs/library/usermanuals/public/29456-d68866-pd-l1-28-8-gastric-interpretation-manual-en-eu.pdf.
- 21. Nuti S, Zhang Y, Zerrouki N, et al. High interobserver and intraobserver reproducibility among pathologists assessing PD-L1 CPS across multiple indications. Histopathology 2022; 81: 732-41. ArticlePubMedPDF
- 22. Park Y, Koh J, Na HY, et al. PD-L1 testing in gastric cancer by the combined positive score of the 22C3 PharmDx and SP263 assay with clinically relevant cut-offs. Cancer Res Treat 2020; 52: 661-70. ArticlePubMedPMCPDF
- 23. Tsao MS, Kerr KM, Kockx M, et al. PD-L1 immunohistochemistry comparability study in real-life clinical samples: results of Blueprint Phase 2 Project. J Thorac Oncol 2018; 13: 1302-11. ArticlePubMedPMC
- 24. Prince EA, Sanzari JK, Pandya D, Huron D, Edwards R. Analytical concordance of PD-L1 assays utilizing antibodies from FDA-approved diagnostics in advanced cancers: a systematic literature review. JCO Precis Oncol 2021; 5: 953-73. ArticlePubMed
- 25. Akhtar M, Rashid S, Al-Bozom IA. PD-L1 immunostaining: what pathologists need to know. Diagn Pathol 2021; 16: 94.ArticlePubMedPMCPDF
- 26. O’Malley DP, Yang Y, Boisot S, et al. Immunohistochemical detection of PD-L1 among diverse human neoplasms in a reference laboratory: observations based upon 62,896 cases. Mod Pathol 2019; 32: 929-42. ArticlePubMedPMCPDF
REFERENCES
Figure & Data
References
Citations

- Adjuvant immunotherapy in patients with resected gastric and oesophagogastric junction cancer following preoperative chemotherapy with high risk for recurrence (ypN+ and/or R1): European Organisation of Research and Treatment of Cancer (EORTC) 1707 VESTIG
F. Lordick, M.E. Mauer, G. Stocker, C.A. Cella, I. Ben-Aharon, G. Piessen, L. Wyrwicz, G. Al-Haidari, T. Fleitas-Kanonnikoff, V. Boige, R. Lordick Obermannová, U.M. Martens, C. Gomez-Martin, P. Thuss-Patience, V. Arrazubi, A. Avallone, K.K. Shiu, P. Artru
Annals of Oncology.2025; 36(2): 197. CrossRef - PD-L1 as a Biomarker in Gastric Cancer Immunotherapy
Yunjoo Cho, Soomin Ahn, Kyoung-Mee Kim
Journal of Gastric Cancer.2025; 25(1): 177. CrossRef - PD-L1 importance in malignancies comprehensive insights into the role of PD-L1 in malignancies: from molecular mechanisms to therapeutic opportunities
Mojdeh Soltani, Mohammad Abbaszadeh, Hamed Fouladseresht, Mark J. M. Sullman, Nahid Eskandari
Clinical and Experimental Medicine.2025;[Epub] CrossRef - CLDN18.2 expression in gastroesophageal adenocarcinoma: prevalence, heterogeneity, and prognostic implications in Spanish patients
Carolina Martinez-Ciarpaglini, María Ortega, Sandra Pérez-Buira, Aitana Bolea, Beatriz Casado Guerra, Carmen Herencia Bellido, Paula Tornero Piñero, Dolores Naranjo-Hans, Brenda Palomar, Hernán Quiceno, Amanda Sardón Fernández, Ariadna Torner Calvo, Feder
Virchows Archiv.2025;[Epub] CrossRef - Distinct clinicopathological and survival profiles of CLDN18.2 and PD-L1 expression in advanced gastric cancer and gastroesophageal junction adenocarcinoma
D.R. Castillo, M. Guo, P. Shah, M. Hazeltin, D. Tai, F. Al-Manaseer, S. Mlamba, D. Perez, S. Yeremian, S. Guzman, R. Mannan, C. Crook, C. Lau, N. Tawar, G. Brar, M. Raoof, Y. Woo, S.P. Wu, D. Li
ESMO Gastrointestinal Oncology.2025; 10: 100261. CrossRef - Best Practice PD-L1 Staining and Interpretation in Gastric Cancer Using PD-L1 IHC PharmDx 22C3 and PD-L1 IHC PharmDx 28-8 Assays, with Reference to Common Issues and Solutions
Soomin Ahn, Inwoo Hwang, Yuyeon Kim, Somin Lee, Yunjoo Cho, So Young Kang, Deok Geun Kim, Jeeyun Lee, Kyoung-Mee Kim
Biomedicines.2025; 13(11): 2824. CrossRef - PD-L1 thresholds predict efficacy of immune checkpoint inhibition in first-line treatment of advanced gastroesophageal adenocarcinoma. A systematic review and meta-analysis of seven phase III randomized trials
V. Formica, C. Morelli, L. Fornaro, S. Riondino, M. Rofei, E. Fontana, E.C. Smyth, M. Roselli, H.-T. Arkenau
ESMO Open.2024; 9(11): 103967. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure



















Fig. 1.
Fig. 2.
Fig. 3.
Fig. 4.
Fig. 5.
Fig. 6.
Fig. 7.
Fig. 8.
Fig. 9.
Fig. 10.
Fig. 11.
Fig. 12.
Fig. 13.
Fig. 14.
Fig. 15.
Fig. 16.
Fig. 17.
Fig. 18.
Fig. 19.
| CheckMate-649 | KEYNOTE-811 | KEYNOTE 859 | RATIONALE-305 | |
|---|---|---|---|---|
| Candidates | HER2-negative GC | HER2-positive GC | HER2-negative GC | HER2-negative GC |
| Drug | Nivolumab | Pembrolizumab | Pembrolizumab | Tislelizumab |
| PD-L1 assay | 28-8 pharmDx | 22C3 pharmDx | 22C3 pharmDx | SP263 |
| Antibody supplier | Dako (Agilent) | Dako (Agilent) | Dako (Agilent) | Ventana (Roche) |
| Antibody species | Rabbit mAb | Mouse mAb | Mouse mAb | Rabbit mAb |
| Scoring | CPS | CPS | CPS | TAP |
| Cutoff | 5 | 1 | TBD | 5% |
| US Food and Drug Administration | Approved (on Apr 16, 2021) | Approved (on Aug 29, 2023) | Approved (on Nov 16, 2023) | Not yet |
| Korean Ministry of Food and Drug | Approved (on Sep 1, 2023 | Approved (on Dec 19, 2023) | Not yet | Not yet |
| PD-L1 assay | Cutoff | No. of observers | No. of cases | Sample type | Interobserver agreement | Fleiss kappa value | Reference |
|---|---|---|---|---|---|---|---|
| 22C3 PharmDx | CPS ≥ 1 | 3 | 68 | Not mentioned | OPA 96.6% | - | [13] |
| 22C3 PharmDx | CPS ≥ 1 | 120 | 20 (day1), 25 (day2) | Resection | OPA 90.6% | 0.828 | [21] |
| 22C3 PharmDx | CPS value | 5 | 55 | Tissue microarray | ICC 0.387 (lower 95% CI, 20.9%) | CPS ≥ 1 0.389 | [22] |
| SP263 | CPS value | 5 | 55 | Tissue microarray | ICC 0.349 (lower 95% CI, 13.5%) | CPS ≥ 1 0.256 | |
| 22C3 PharmDx | CPS ≥ 1 | 14 | 112 | Biopsy | OPA 31.48% (95% CI, 22.72–40.24) | 0.477 | [18] |
| ICC 0.484 (95% CI, 0.403–0.571) | - | ||||||
| CPS ≥ 10 | 14 | 112 | Biopsy | OPA 67.59% (95% CI, 58.77–76.42) | 0.607 | ||
| ICC 0.604 (95% CI, 0.584–0.624) | - | ||||||
| CPS ≥ 20 | 14 | 112 | Biopsy | OPA 83.33% (95% CI, 76.3–90.36) | 0.626 | ||
| ICC 0.629 (95% CI, 0.562–0.698) | - | ||||||
| 28-8 PharmDx | CPS value | 12 | 100 | Biopsy | ICC 0.45 (95% CI, 0.38–0.53) | - | [19] |
| 22C3 PharmDx | CPS value | 12 | 100 | Biopsy | ICC 0.55 (95% CI, 0.47–0.63) | - |
| Case No. | Institution | Vendor/file format | Specimen type | Interobserver agreement | Consensus CPS |
|---|---|---|---|---|---|
| 1 | A | Philips/.isyntax | Resection | Good |
0 |
| 2 | A | Philips/.isyntax | Biopsy | Poor | 2 |
| 3 | A | Philips/.isyntax | Biopsy | Poor | Not determined |
| 4 | A | Philips/.isyntax | Resection | Poor | 5 |
| 5 | A | Philips/.isyntax | Resection | Good | 5 |
| 6 | A | Philips/.isyntax | Biopsy | Poor | 3 |
| 7 | B | Leica Biosystems/.svs | Biopsy | Poor | 2 |
| 8 | B | Leica Biosystems/.svs | Biopsy | Poor | 5 |
| 9 | B | Leica Biosystems/.svs | Biopsy | Poor | 2 |
| 10 | C | Leica Biosystems/.svs | Biopsy | Good | ≥ 5 |
| 11 | C | Leica Biosystems/.svs | Biopsy | Poor | 3 |
| 12 | C | Leica Biosystems/.svs | Biopsy | Good | ≥ 5 |
| 13 | D | Philips/.isyntax | Resection | Poor | 5 |
| 14 | D | Philips/.isyntax | Biopsy | Poor | 5 |
| 15 | D | Philips/.isyntax | Biopsy | Good | 2 |
| 16 | D | Philips/.isyntax | Resection | Poor | 5 |
| 17 | D | Philips/.isyntax | Biopsy | Poor | 2 |
| 18 | E | 3DHistech/.MRXS | Biopsy | Good | 5 |
| 19 | F | Philips/.isyntax | Biopsy | Good | ≥ 5 |
| 20 | G | Leica Biosystems/.svs | Resection | Poor | 0 |
PD-L1, programmed death-ligand 1; HER2, human epidermal growth factor receptor 2; GC, gastric cancer; CPS, combined positive score; TBD, to be determined; TAP, tumor area positivity.
PD-L1, programmed death-ligand 1; CPS, combined positive score; OPA, overall percentage agreement; ICC, intraclass correlation coefficient; CI, confidence interval.
CPS, combined positive score. Interobserver agreement was defined as good when more than 80% of pathologists agreed on CPS 5 cutoff.

 E-submission
E-submission