Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 46(2); 2012 > Article
-
Original Article
Difference of Genome-Wide Copy Number Alterations between High-Grade Squamous Intraepithelial Lesions and Squamous Cell Carcinomas of the Uterine Cervix - Bum Hee Lee,, Sangyoung Roh1,, Yu Im Kim, Ahwon Lee2, Su Young Kim
-
Korean Journal of Pathology 2012;46(2):123-130.
DOI: https://doi.org/10.4132/KoreanJPathol.2012.46.2.123
Published online: April 25, 2012
Department of Pathology, The Catholic University of Korea School of Medicine, Seoul, Korea.
1Department of Internal Medicine, The Catholic University of Korea School of Medicine, Seoul, Korea.
2Department of Hospital Pathology, The Catholic University of Korea School of Medicine, Seoul, Korea.
- Corresponding Author: Su Young Kim, M.D. Department of Pathology, The Catholic University of Korea, College of Medicine, 222 Banpo-daero, Seocho-gu, Seoul 137-701, Korea. Tel: +82-2-2258-7315, Fax: +82-2-537-6586, suyoung@catholic.ac.kr
- *Bum Hee Lee and Sangyoung Roh contributed equally to this work.
© 2012 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- About 10% of high-grade squamous intraepithelial lesions (HSILs) progress to invasive carcinomas within 2-10 years. By delineating the events that occur in the early stage of the invasion, the pathogenesis of cervical cancer could be better understood. This will also propose the possible methods for inhibiting the tumor invasion and improving the survival of patients.
-
Methods
- We compared the genomic profiles between the HSIL and the invasive squamous cell carcinoma (SCC) using an array comparative genomic hybridization. Using recurrently altered genes, we performed a principal component analysis to see variation of samples in both groups. To find possibly affected pathways by altered genes, we analyzed genomic profiles with the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database and GOEAST software.
-
Results
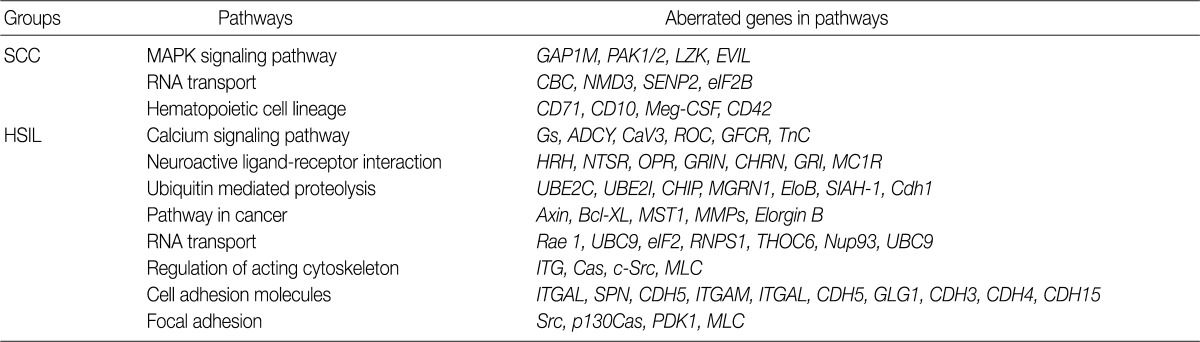
- We found 11q12.3 and 2p24.1 regions have recurrent copy number gains in both groups. 16p12-13 and 20q11-13 regions showed an increased copy number only in cases of HSIL. 1q25.3 and 3q23-29 regions showed copy number gains only in cases of SCC. Altered genes in the SCC group were related to the mitogen-activated protein kinase signaling pathway and the RNA transport. Altered genes in the HSIL group were related to the ubiquitin mediated proteolysis and cell adhesion molecules.
-
Conclusions
- Our results showed not only that gains in 11q12.3 and 2p24.1 were early events occurring in the premalignant lesions and then maintained in cases of SCC but also that gains in 1q25.3 and 3q23-29 were late events occurring after invasion in those of SCC.
- Primary tumor tissues
- In eight cases of uterine cervix tissues, the tissue samples were fixed in a formalin solution and then prepared into paraffin-embedded sections (FFPE). The protocol of the current study was approved by the Institutional Review Board (IRB) (CUMC10U917) of the Catholic University of Korea, College of Medicine. These eight cases consisted of four cases of HSIL and another four cases of invasive SCC (Table 1).
- Genomic DNA extraction
- Unstained tissue sections were cut from the paraffin blocks and then deparaffinized with xylene twice. Normal areas of cervical tissues were removed from the sections using knife blades. The remaining tissues were collected in 1.5 mL microtubes and then digested for 72 hours in the dry chamber, where the temperature was set at 58℃, using Proteinase K (Invitrogen, Carlsbad, CA, USA) in ATL buffer (Qiagen, Valencia, CA, USA) with overlaid mineral oil (Sigma-Aldrich, St. Louis, MO, USA) that was applied to hinder the evaporation and concentration of the buffer. After the tissue digestion, genomic DNAs were extracted using DNeasy blood and tissue kit (Qiagen) according to the manufacturer's protocol. The concentration of the DNA was measured using NanoDrop (Thermo Scientific, Wilmington, DE, USA) and the integrity of the DNA was checked with a 1% agarose gel electrophoresis.
- DNA fragmentation
- Usually the genomic DNA extracted from FFPE tissue is partially degraded and chemically modified.22 The FFPE DNA produces a high-level of noise with standard array CGH protocol. To minimize the level of noise in an array CGH data, random fragmentation method using Shrimp Recombinant DNase (Affymetrix, Santa Clara, CA, USA) was exploited.22 Briefly, a 2.5 µg of extracted DNA and a 1 µg of reference DNA (Promega, Madison, WI, USA) were digested with the Shrimp Recombinant DNase (Affymetrix) to get DNA fragments of 200 to 400 bp in size.
- Array CGH
- Digested fragments of extracted DNA and reference DNA were labeled with Cyanine-5 and Cyanine-3 dUTP, respectively, using the Bioprime array CGH genomic labeling system (Invitrogen). After labeling reaction, the amount of labeled product and the efficiency of labeling were measured using Nanodrop (Thermo Scientific). Each labeled DNA was co-hybridized at an amount of 4 µg, identical to that of labeled reference DNA, to the 4×44K human aCGH array platform (Agilent Technologies, Santa Clara, CA, USA). Hybridization was performed at 65℃, for which the array chamber was rotated at 20 rpm for 24 hours. Then the arrays were washed according to the manufacturer's instructions. Microarray slides were scanned on a GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA, USA). The scanned images were analyzed using Feature Extraction software (Agilent Technologies).
- Microarray data analysis
- To define segments with an altered copy number, we used the aberration detection method 2 (ADM-2) algorithm with a threshold of 6 using DNA analytics software (Agilent Technologies).23 To minimize false positives, we excluded aberrations spanning less than three consecutive probes from further analysis. To focus on recurrent changes in each group, we used aberrations found in at least two cases (50%) of each group. Besides, we used R 2.13.1 software (R Foundation for Statistical Computing, Vienna, Austria) and Matlab 7.12 software (Mathworks, Natick, MA, USA) for statistical calculations and graphical presentations.
- Real-time polymerase chain reaction (PCR) of syndecan1 (SDC1) gene
- Of the samples that showed an increased copy number of SDC1 on an array CGH, three cases were selected to confirm the copy number gain of SDC1 in the cervical lesion. A 20 ng of genomic DNA extracted from the cervical lesion was used for each reaction. SYBR Premix Ex Taq (Takara Bio Inc., Shiga, Japan) was used to amplify target (SDC1) and reference (beta actin, ACTB) genes using a LightCycler 480 II (Roche, Rotkreuz, Switzerland). We designed two pairs of primers for SDC1 to detect copy number of two regions (exon 2 and exon 3) of SDC1 and one pair for ACTB as reference. The following oligonucleotides were used as the primer: SDC1-2F (exon 2) 5'-GTTTTTGCAACGGCTAAGGA-3'; SDC1-2R (exon 2) 5'-TCTACTGCCGGATTCCTCTC-3'; SDC1-3F (exon 3) 5'-CGGCCAAGCCTTTACTCATA-3'; SDC1-3R (exon 3) 5'-AGCCGGAGAAGTTGTCAGAG-3'; ACTB-F 5'-AGAAAATCTGGCACCACACC-3'; ACTB-R 5'-AACGGCAGAAGAGAGAACCA-3'.
- After the pre-incubation at 95℃ for 5 minutes, 45 cycles of the PCR (95℃ for 30 seconds, 60℃ for 30 seconds, and 72℃ for 30 seconds) were performed. Then, the melting curves were analyzed to confirm specific PCRs; all samples were performed in triplicate. We used pooled genomic DNA extracted from the stromal tissues in the same cases as a calibrator. We used the E-method of LightCycler 480 II (Roche) to calculate template concentration, where we normalized the copy number of the target and reference using the calibrator.
- Pathway analysis
- To find possibly affected pathways consisting of altered genes in cases of cervical cancer, we searched the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (http://www.genome.jp/kegg/pathway.html).
- Gene ontology (GO) analysis
- To analyze a set of genes with commonly altered copy numbers based on a gene ontology, we used Gene Ontology enrichment analysis software toolkit (GOEAST).24 The list of gene symbols showing a recurrent aberration of the copy number was submitted to the GOEAST. Of the GO term IDs that the GOEAST identified from the list, those with p-value of <0.05 were considered to be statistically significant.
MATERIALS AND METHODS
- Distribution of the alteration of copy number
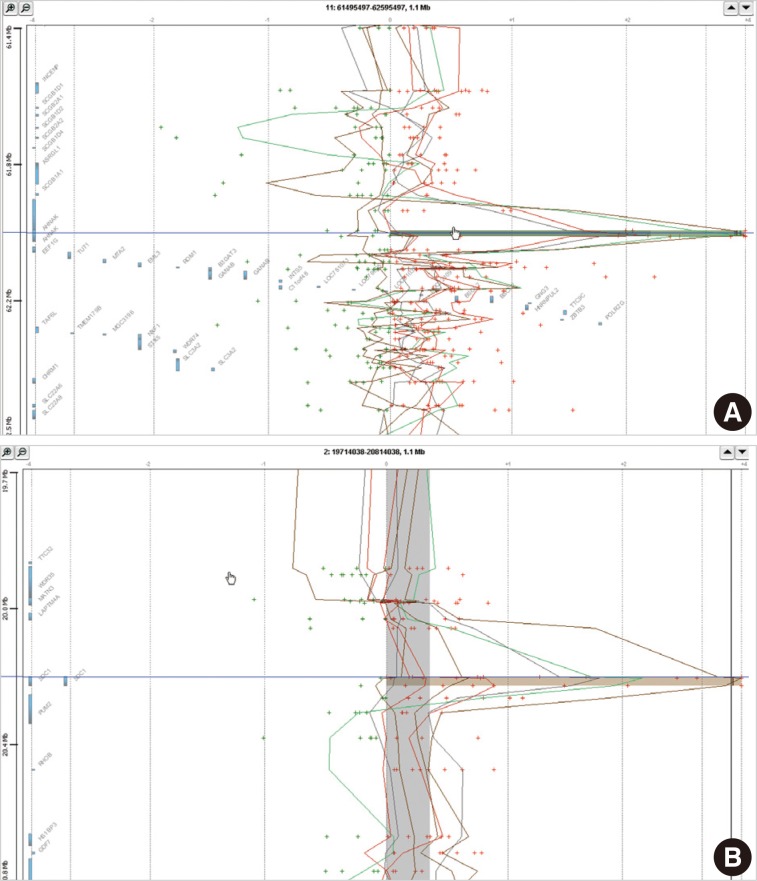
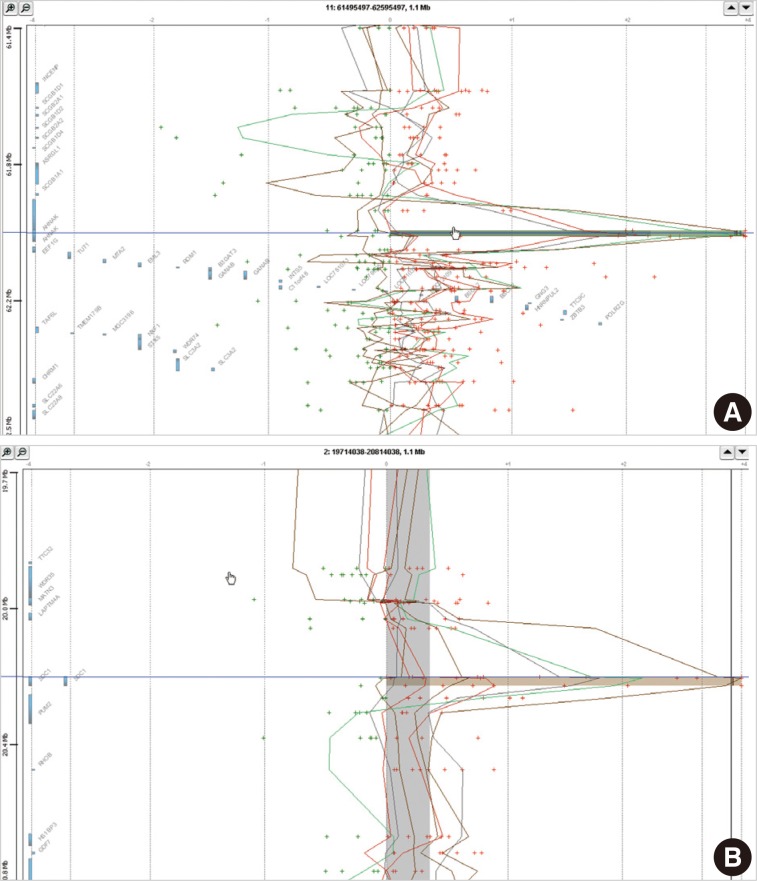
- In eight cases of cervical cancer, we found 18,472 microarray probes showing the alteration of copy number. To find recurrent aberrations in each group, we collected aberrations detected in at least two cases of each group (Table 2). No regions showed a decreased copy number in two or more cases in each group. 11q12.3 (AHNAK, TRIM29) and 2p24.1 (SDC1) were found to show a copy number gain in both the HSIL group and the SCC group (Fig. 1). The recurrent aberrations of the 16p12-13 and 20q11-13 regions were found only in the HSIL group and those of the 1q25.3 (IVNS1SBP) and 3q23-29 regions were found only in the SCC group.
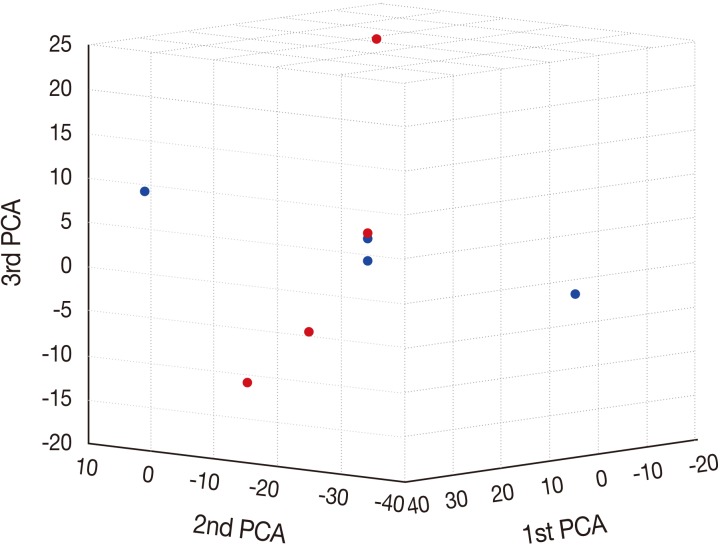
- Principal component analysis (PCA)
- To characterize the aberration profiles of the tissue samples of HSIL and SCC, we performed the PCA using all aberration information (Fig. 2). There was a variability in the first, second and third factors forming the principal components, approximately showing an orthogonal linear relationship between HSIL and SCC. This indicates that there is a distinct difference in the pattern of the variation between HSIL and SCC based on their genomic profile.
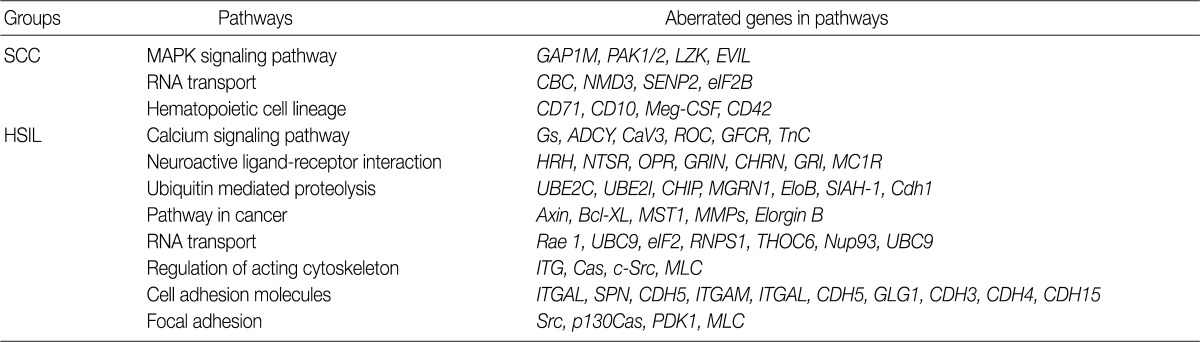
- Analysis of the KEGG pathway database
- To find possibly affected pathways consisting of altered genes in cases of cervical cancer, we searched the KEGG pathway database (Table 3). The mitogen-activated protein kinase signaling pathway is found in genes with an aberration of the copy number in cases of SCC. In cases of HSIL with an alteration of the copy number, pathways related to ubiquitin-mediated proteolysis and cell adhesion molecules were found. RNA transport pathway was found in both groups.
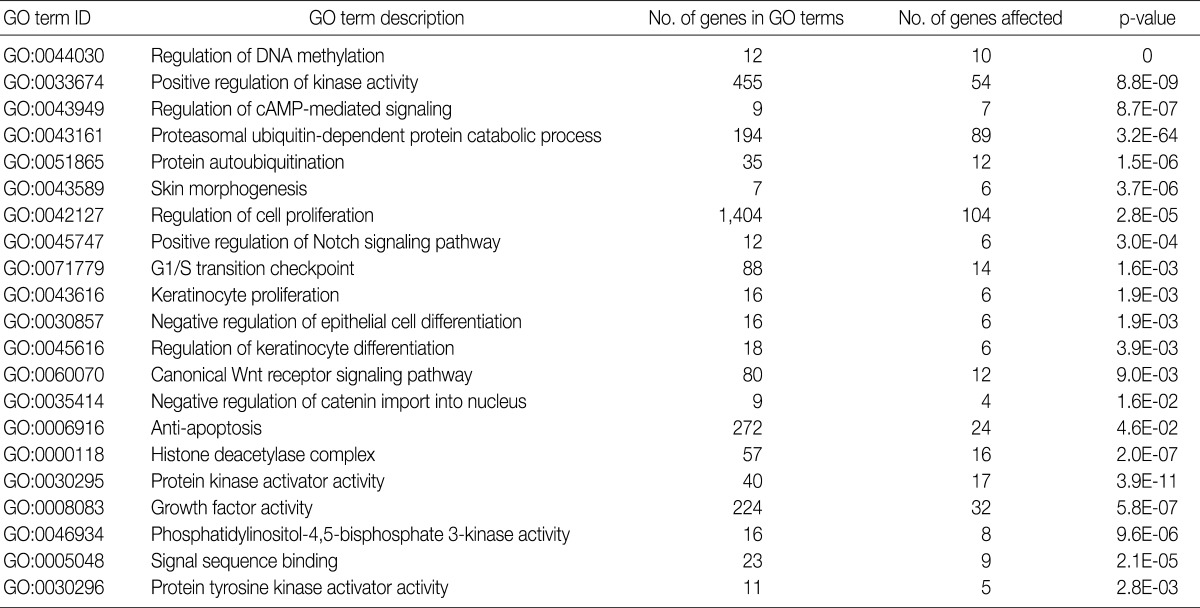
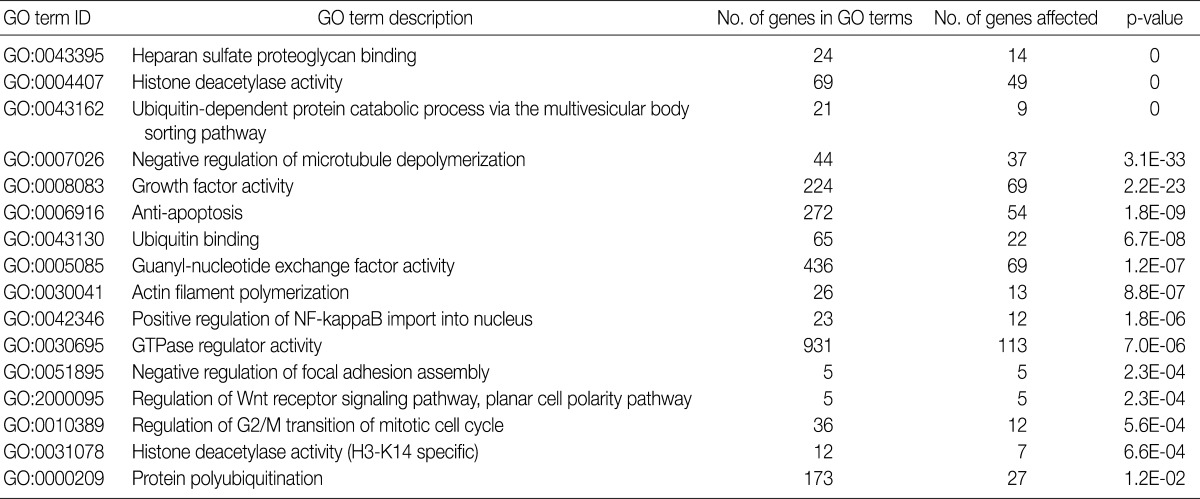
- GO analysis
- Using the GOEAST, we found 757 significant GO term IDs from the list of genes with an alteration of the copy number only in the SCC group. We also found 407 GO term IDs from the list of genes with an alteration of the copy number in both groups. Of the GO term IDs that had been found, representative cancer-related IDs are represented in Tables 4 and 5.
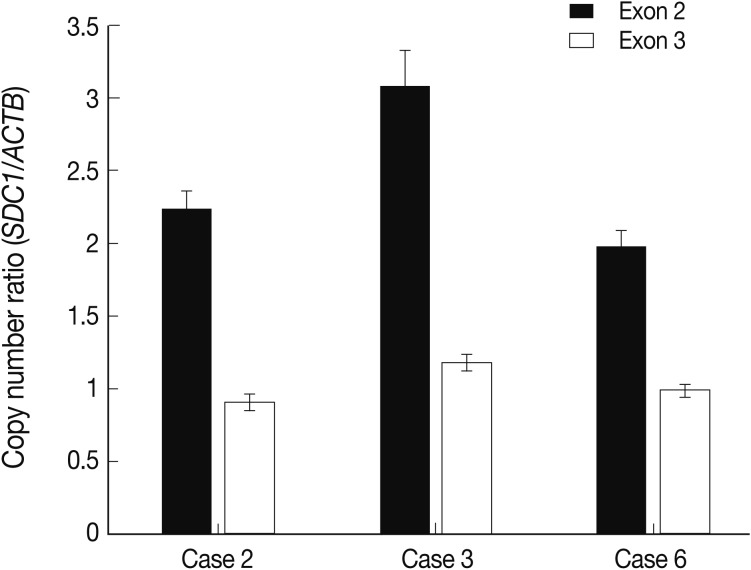
- The alteration of copy number of SDC1 gene
- Of the genes with an aberration of the copy number, we were interested in SDC1 whose gene product is involved in the cell binding, cytoskeletal organization and might play a role in the tumor invasion and metastasis of the tumor. To confirm the dose of an alteration of the copy number in SDC1 gene, we performed the real-time PCR. There three cases of an increased copy number in the region of exon 2 but not in that of exon 3 (Fig. 3).
RESULTS
- In the current study, we screened whole genomes with any recurrent alterations of the copy number in cases of cervical SCC. We also analyzed a set of genes with an alteration of the copy number using bioinformatics database and tools. Our results were partly in agreement with previous reports about cervical cancer. Of the alterations of the copy number that have been reported up to present, gains of 1q and 3q are frequently mentioned on a variety of reports about cervical cancer.3,11,12 Our results showed that the gain of 3q was a pivotal aberration during the transition from HSIL to invasive SCC.21 Using an array CGH, we narrowed down the region to 3q23-29 and thereby detected another recurrent aberration of IVNS1ABP during the above transition. By contrast, a loss of the copy number has not been frequently reported on the articles. We could not also identify any recurrent losses of the copy number in the current study. This suggests that a gain of the copy number rather than a loss of it might be more relevant in the pathogenesis of cervical cancer.
- Advanced cancers are known to have chromosomal instabilities. Due to the instability, cancer genomes harbor many non-specific patterns of the alteration of copy number. By focusing on relatively earlier stage of SCC (International Federation of Genycology and Obstetrics [FIGO] stage IIb) and comparing its findings with those of premalignant lesions (HSIL), we were successful in not only demonstrating that there was a difference in the alteration between SCC and HSIL but also minimizing nonspecific patterns of the alterations due to the chromosomal instability.
- In cancer cells cultured in in vitro conditions, there is a tendency to randomly accumulate the alteration of copy number due to chromosomal instabilities.25 Cell lines, derived from malignant cells and grown in culture condition, harbor many nonspecific patterns of the alteration of copy number. This is seen after the collection of tissue samples from patients. It is therefore probable that reports about the alterations of copy number using cell lines may not be applicable to cancer cells in an in vivo condition. To minimize this bias of cell line study, we used primary malignant and premalignant tissues to investigate the alteration of copy number.
- Of the alteration of copy number that had been reported from cervical cancer, some had a prognostic value. Oncostatin M receptor (OSMR) frequently shows a copy number gain in cervical cancer, which is associated with adverse clinical outcomes.26 Our results showed, however, that there was no recurrent aberration of the copy number in the 5p13.1 region where OSMR is located. This discrepancy can be explained by OSMR gain is a late event in cervical carcinogenesis as the authors26 mentioned. In their report, more than half of the samples they studied were of FIGO stage IIIb or higher.
- Another variant of cervical cancer, cervical adenocarcinoma shows different patterns of the alteration of copy number from SCC. It has been reported that cervical adenocarcinomas have frequent gains at 3q, 17q, 1p, 1q, and 11q.27 According to these authors,27 the gain was more commonly seen than the loss of copy number. This was also shown in the current study. In addition, 3q28-29 gain was found in both the above authors' reports and our findings.
- It is not always observed that the alteration of copy number results in that of gene expression. In about half of total cases, the alteration of the copy number leads to that of the gene expression. According to the combination study about the dosage and expression of genes, EPHB2, MMP1, AKT1, ABCC3 genes are both amplified and over-expressed in cases of cervical cancer.28
- To analyze the possible net effect of genomic alterations, we searched for the KEGG pathway database with lists of genes with an alteration of the copy number. In cases of HSIL, genes with an alteration of the copy number are related to both the pathways of ubiquitin-mediated proteolysis and adhesion related pathways (Table 3). HPV E6 induces the rapid degradation of p53 via ubiquitin-dependent proteolysis.1 In cases of HSIL, the recurrent copy number gain of genes related to ubiquitin-mediated proteolysis might explain the possible underlying mechanisms by which the rapid degradation of p53 in HPV-infected cells occurs before entry into the invasive stage of SCC.
- There are no reports explaining the reason why there was no persistent presence of the alteration of copy number in cases of HSIL. This might be due to chromosomal instability in malignant cells.
- We have previously reported an increased expression of SDC1 in cases of cervical cancer and its association with a prolonged survival.29 In the previous report, we performed an interphase FISH to investigate the alteration of copy number of SDC1 but in vain. The detection limit of FISH is generally set at 10 Mbp. Conventional types of the FISH analysis could not therefore detect any changes with a span of <10 Mbp. If the alteration of copy number of SDC1 gene is partly present, as we shown in the current study, it might not be detected on the FISH analysis. This explains the discrepancy between our previous report and the current study.
- Limitations of the current study are due to a relatively smaller number of samples. In the current study, we used only four samples of HSIL and another four samples of invasive SCC. Due to a smaller number of samples, the nature of genome-wide alteration of copy number may not be fully explained in cases of HSIL and those of invasive SCC. Nevertheless, we applied stringent methods to minimize false positives and get pertinent findings in a total of eight cases of samples. In addition, we have intentionally excluded any alterations with a span of <three consecutive probes on the genome from the data analysis to resolve two problems. The one problem arises from the FFPE DNA samples. Modified genomic DNA from the FFPE tissues can over-represent or under-represent the original copy number of genomic DNA on certain probes. The other problem arises from the probes on microarrays. The probes are well designed and their applicability has been validated by a wide range of researchers, although one or two misbehaving probes may show erroneous results. By ruling out the results obtained from one or two consecutive probes, we resolved both problems mentioned above. It is unavoidable, however, that this strategy increases false negatives. This might be partly because we found no recurrent loss of the copy number in the current study.
- In the current study, we used HSIL and SCC tissues obtained from different patients. If we had compared the tissue samples of HSIL and SCC obtained from the same patient, we could have better shown the direct relationship between the two lesions. In the methods of the current study, we had required a significant amount of genomic DNA with a good quality. But we could not obtained a sufficient amount of genomic DNA from the same patient for an array CGH, the real-time PCR and other necessary procedures for quality control. We collected tissue samples of HSIL and SCC from the different patients, but tried to find recurrent changes in both groups. To our knowledge, an analysis of the recurrent alterations will reduce a bias due to the difference between the individual cases and it will also provide the information about the role of the alteration of copy number that can be generally accepted as the pathogenesis of cervical cancer.
- To summarize, our results showed that the recurrent alteration of copy number occurred in cases of HSIL and those of SCC based on the genomic profiles of both disease entities. The alteration of copy number in AHNAK, TRIM29, and SDC1 genes and that in IVNS1ABP gene were found in premalignant lesions and invasive SCC, respectively.
DISCUSSION
Acknowledgments
Acknowledgments
- 1. Robbins SL, Kumar V, Abbas AK, Cotran RS, Fausto N. Robbins and Cotran pathologic basis of disease. 2010; Philadelphia: Saunders.
- 2. de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology 2004; 324: 17-27. ArticlePubMed
- 3. Wilting SM, de Wilde J, Meijer CJ, et al. Integrated genomic and transcriptional profiling identifies chromosomal loci with altered gene expression in cervical cancer. Genes Chromosomes Cancer 2008; 47: 890-905. ArticlePubMedPMC
- 4. Castro FA, Ivansson EL, Schmitt M, et al. Contribution of TMC6 and TMC8 (EVER1 and EVER2) variants to cervical cancer susceptibility. Int J Cancer 2012; 130: 349-355. ArticlePubMedPMC
- 5. Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature 1998; 396: 643-649. ArticlePubMedPDF
- 6. Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235: 177-182. ArticlePubMed
- 7. Han CB, Ma JT, Li F, et al. EGFR and KRAS mutations and altered c-Met gene copy numbers in primary non-small cell lung cancer and associated stage N2 lymph node-metastasis. Cancer Lett 2012; 314: 63-72. ArticlePubMed
- 8. Cheng I, Levin AM, Tai YC, et al. Copy number alterations in prostate tumors and disease aggressiveness. Genes Chromosomes Cancer 2012; 51: 66-76. ArticlePubMedPMC
- 9. Rømer MU, Jensen NF, Nielsen SL, et al. TOP1 gene copy numbers in colorectal cancer samples and cell lines and their association to in vitro drug sensitivity. Scand J Gastroenterol 2012; 47: 68-79. ArticlePubMed
- 10. Stankiewicz P, Beaudet AL. Use of array CGH in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr Opin Genet Dev 2007; 17: 182-192. ArticlePubMed
- 11. Wilting SM, Snijders PJ, Meijer GA, et al. Increased gene copy numbers at chromosome 20q are frequent in both squamous cell carcinomas and adenocarcinomas of the cervix. J Pathol 2006; 209: 220-230. ArticlePubMed
- 12. Halder A, Halder S, Fauzdar A. A preliminary investigation of genomic screening in cervical carcinoma by comparative genomic hybridization. Indian J Med Res 2005; 122: 434-446. PubMed
- 13. Hidalgo A, Baudis M, Petersen I, et al. Microarray comparative genomic hybridization detection of chromosomal imbalances in uterine cervix carcinoma. BMC Cancer 2005; 5: 77.ArticlePubMedPMCPDF
- 14. Rao PH, Arias-Pulido H, Lu XY, et al. Chromosomal amplifications, 3q gain and deletions of 2q33-q37 are the frequent genetic changes in cervical carcinoma. BMC Cancer 2004; 4: 5.ArticlePubMedPMCPDF
- 15. Umayahara K, Numa F, Suehiro Y, et al. Comparative genomic hybridization detects genetic alterations during early stages of cervical cancer progression. Genes Chromosomes Cancer 2002; 33: 98-102. ArticlePubMed
- 16. Hidalgo A, Schewe C, Petersen S, et al. Human papillomavirus status and chromosomal imbalances in primary cervical carcinomas and tumour cell lines. Eur J Cancer 2000; 36: 542-548. ArticlePubMed
- 17. Heselmeyer K, Macville M, Schröck E, et al. Advanced-stage cervical carcinomas are defined by a recurrent pattern of chromosomal aberrations revealing high genetic instability and a consistent gain of chromosome arm 3q. Genes Chromosomes Cancer 1997; 19: 233-240. ArticlePubMed
- 18. Allen DG, White DJ, Hutchins AM, et al. Progressive genetic aberrations detected by comparative genomic hybridization in squamous cell cervical cancer. Br J Cancer 2000; 83: 1659-1663. ArticlePubMedPMC
- 19. Cooke SL, Temple J, Macarthur S, et al. Intra-tumour genetic heterogeneity and poor chemoradiotherapy response in cervical cancer. Br J Cancer 2011; 104: 361-368. ArticlePubMedPMCPDF
- 20. Dellas A, Torhorst J, Jiang F, et al. Prognostic value of genomic alterations in invasive cervical squamous cell carcinoma of clinical stage IB detected by comparative genomic hybridization. Cancer Res 1999; 59: 3475-3479. PubMed
- 21. Heselmeyer K, Schröck E, du Manoir S, et al. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci U S A 1996; 93: 479-484. ArticlePubMedPMC
- 22. Hostetter G, Kim SY, Savage S, et al. Random DNA fragmentation allows detection of single-copy, single-exon alterations of copy number by oligonucleotide array CGH in clinical FFPE samples. Nucleic Acids Res 2010; 38: e9.ArticlePubMedPMC
- 23. Przybytkowski E, Ferrario C, Basik M. The use of ultra-dense array CGH analysis for the discovery of micro-copy number alterations and gene fusions in the cancer genome. BMC Med Genomics 2011; 4: 16.ArticlePubMedPMCPDF
- 24. Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res 2008; 36: W358-W363. ArticlePubMedPMC
- 25. Pihan GA, Purohit A, Wallace J, et al. Centrosome defects and genetic instability in malignant tumors. Cancer Res 1998; 58: 3974-3985. PubMed
- 26. Ng G, Winder D, Muralidhar B, et al. Gain and overexpression of the oncostatin M receptor occur frequently in cervical squamous cell carcinoma and are associated with adverse clinical outcome. J Pathol 2007; 212: 325-334. ArticlePubMed
- 27. Yang YC, Shyong WY, Chang MS, et al. Frequent gain of copy number on the long arm of chromosome 3 in human cervical adenocarcinoma. Cancer Genet Cytogenet 2001; 131: 48-53. ArticlePubMed
- 28. Narayan G, Bourdon V, Chaganti S, et al. Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: identification of candidate amplified and overexpressed genes. Genes Chromosomes Cancer 2007; 46: 373-384. ArticlePubMed
- 29. Kim YI, Lee A, Lee BH, Kim SY. Prognostic significance of syndecan-1 expression in cervical cancers. J Gynecol Oncol 2011; 22: 161-167. ArticlePubMedPMC
REFERENCES

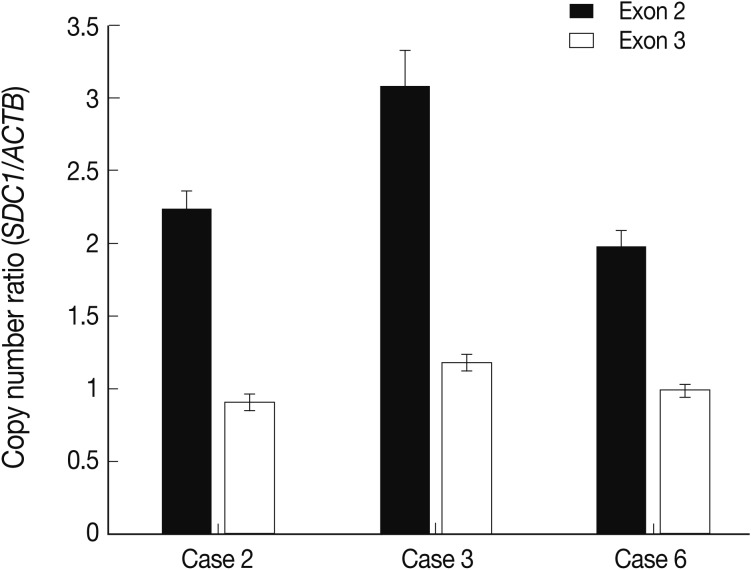
Figure & Data
References
Citations

- Cytokeratin and protein expression patterns in squamous cell carcinoma of the oral cavity provide evidence for two distinct pathogenetic pathways
GESCHE FROHWITTER, HORST BUERGER, PAUL J. VAN DIEST, EBERHARD KORSCHING, JOHANNES KLEINHEINZ, THOMAS FILLIES
Oncology Letters.2016; 12(1): 107. CrossRef - 'Drawing' a Molecular Portrait of CIN and Cervical Cancer: a Review of Genome-Wide Molecular Profiling Data
Olga V Kurmyshkina, Pavel I Kovchur, Tatyana O Volkova
Asian Pacific Journal of Cancer Prevention.2015; 16(11): 4477. CrossRef



Fig. 1
Fig. 2
Fig. 3
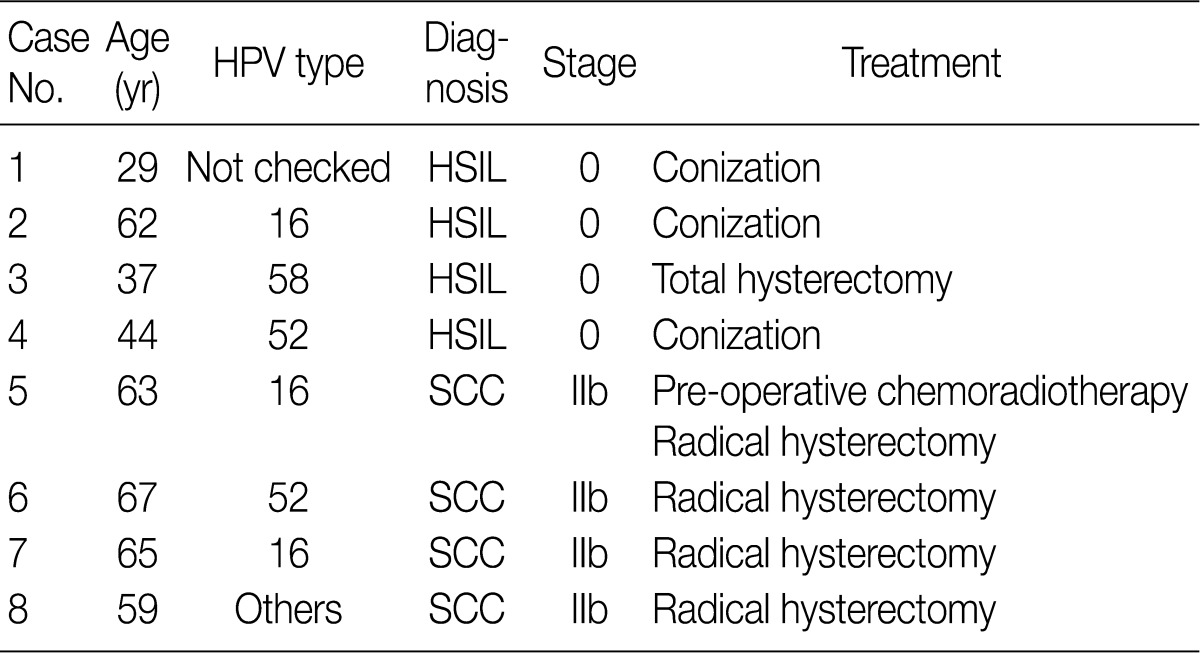



HPV, human papillomavirus; HSIL, high-grade squamous intraepithelial lesion; SCC, squamous cell carcinoma.
HSIL, high-grade squamous intraepithelial lesion; SCC, squamous cell carcinoma.
KEGG, Kyoto Encyclopedia of Genes and Genomes; SCC, squamous cell carcinoma; MAPK, mitogen-activated protein kinase; HSIL, high-grade squamous intraepithelial lesion.
GO, gene ontology; SCC, squamous cell carcinoma.
GO, gene ontology

 E-submission
E-submission

 PubReader
PubReader Cite this Article
Cite this Article