Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 46(2); 2012 > Article
-
Original Article
Markers for Screening Lynch Syndrome Are Reliable and Useful for Identifying the Specimen Mislabeling - Sun-ju Byeon, Jiwoon Choi, Kyung Han Nam, Bo-Gun Jang, Hee Eun Lee, Min A Kim, Woo Ho Kim
-
Korean Journal of Pathology 2012;46(2):131-136.
DOI: https://doi.org/10.4132/KoreanJPathol.2012.46.2.131
Published online: April 25, 2012
Department of Pathology, Seoul National University College of Medicine, Seoul, Korea.
- Corresponding Author: Woo Ho Kim, M.D. Department of Pathology, Seoul National University College of Medicine, 103 Daehak-ro, Jongno-gu, Seoul 110-799, Korea. Tel: +82-2-740-8269, Fax: +82-2-765-5600, woohokim@snu.ac.kr
© 2012 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- During specimen processing in surgical pathology laboratories, specimen-related adverse events (SRAEs), such as mislabeling and specimen mixed-up might occur. In these situations, molecular techniques using short tandem repeat (STR) loci are required to identify the personal identity. Microsatellite instability (MSI) test is widely used for screening the hereditary non-polyposis colon cancer (Lynch syndrome) in surgical pathologies using polymorphic STR markers. We tried to evaluate the applicability of the MSI test for SRAEs.
-
Methods
- We obtained 253 MSI test results to analyze the allele frequencies. After calibrating the estimated nucleotide lengths, we calculated the allele frequencies, a random match probability, and a likelihood ratio (LR) of three dinucleotide STR markers (D5S349, D17S250, and D2S123).
-
Results
- The distribution of LR was 136.38 to 5,606,213.10. There was no case of LR<100. In addition, there were 153 cases (60.5%) of LR ranging from 100 to 10,000 and 100 cases (39.5%) of LR>10,000. Furthermore, the combined probability of identity was 9.23×10-4 and the combined power of exclusion was 0.99908.
-
Conclusions
- Using the three STR markers that are recommended for MSI test, all the cases were positively identified in 1% range and about one-third cases showed high LR (>10,000). These results showed that MSI tests are useful to screen the personal identity in case of SRAE in pathology laboratories.
- MSI test
- To perform MSI test, we differentially marked the tumor and normal mucosa on hematoxylin and eosin staining sections of tissue samples on slide glasses. DNA was harvested from the tissue samples of 10 µm thickness sections which were fixed with either methacarn or formalin and embedded with paraffin after microdissection. Multiplex-polymerase chain reaction (PCR) procedures were performed according to the previously described method (Table 1).9 ABI Prism 3100 Genetic Analyzer (Applied Biosystems) and GeneScan v3.7 software (Applied Biosystems) were used to perform the MSI test.
- Data analysis and calibration
- We sampled 253 cases of MSI test results that had been reported during the period from April to June 2011. Because an MSI analysis of the tumor DNA may show unstable alleles and two mononucleotide repeat STRs (BAT25 and BAT26) are difficult for identifying exact allele (data not shown), we used the results from the three dinucleotide repeat STR loci (D5S349, D17S250, and D2S123) from normal mucosa for allele frequencies analysis. To estimate the length of nucleotide of each allele, we adopted the local southern sizing algorithm.10 Then we corrected the estimated value of the length of nucleotide lengths and round them off using EXCEL 2010 (Microsoft, Redmond, WA, USA) and R v2.14.0.11 We assumed that there was no "off-ladder" allele (the allele that does not fall within 0.5 bp of an allele from the corresponding locus) in 3 microsatellite loci.
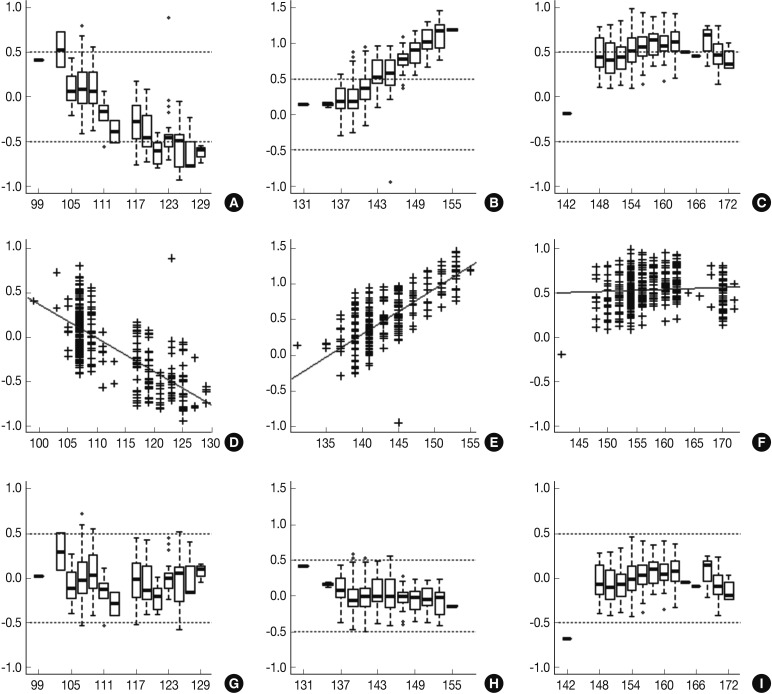
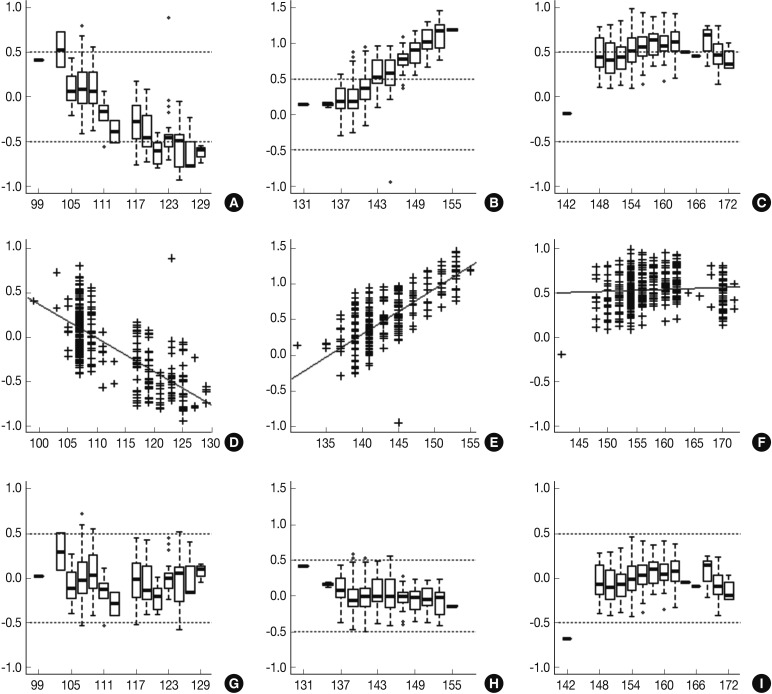
- Following analyses of raw data, there was a gradual change in the difference between the observed nucleotide length (ONL) and the expected nucleotide length (ENL) in each marker. Nine hundred sixty-nine cases of ONL (969/1518, 63.8%) fell within 0.5 bp of the ENL. It was difficult to apply a single algorithm for general regression to calculate the entire ONL. We therefore calculated regression coefficients of each marker using a linear regression method. Calibration formula was followed as:
- Calibrated nucleotide length=ONL-Calculated difference between ENL and ONL=ONL-(Intercept+Slope×ONL)=(1-slope)×ONL-Intercept
- Regression coefficients and statistical parameters of D5S346, D17S250, and D2S123 are represented in Table 2. Raw and calibrated data are shown in Fig. 1. Following the calibration, only 32 cases of ONL (32/1,518, 2.1%) did not fall within 0.5 bp of the ENL.
- Calculation of the random probability and LR
- The calculation of the homozygote and heterozygote frequencies in population structure was proposed by National Research Council II (NRC-II) recommendation 4.1. In a general population (inbreeding coefficient, θ=0.01), homozygote and heterozygote frequency were "pi2+pi (1-pi)θ (pi; allele frequency)" and "2pipj(1-θ) (pi, pj; allele frequency)," respectively. We calculated the LR using a formula: LR=1/RMP.12
MATERIALS AND METHODS
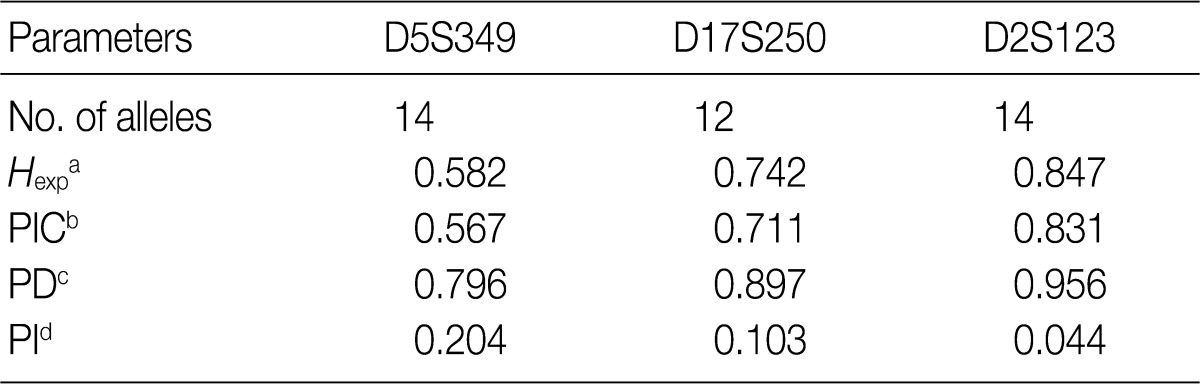
- Allele frequencies and statistical parameters in the three microsatellite loci
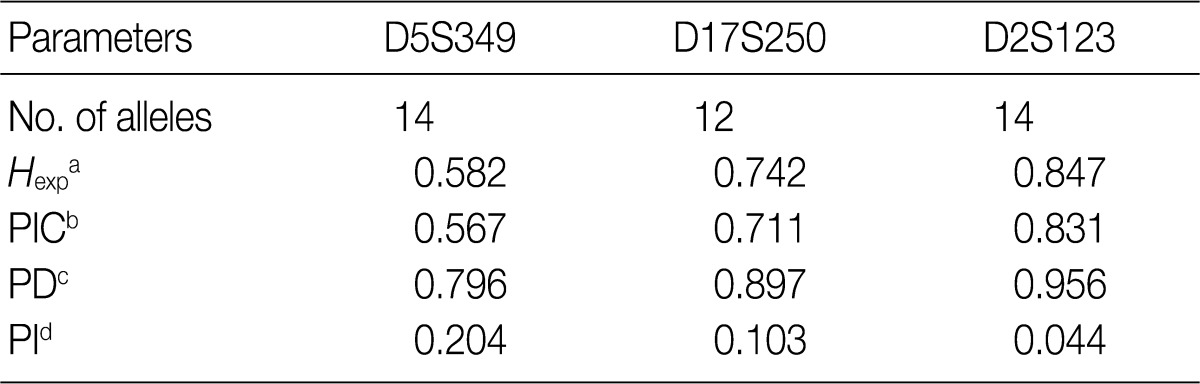
- Allele frequencies and statistical parameters of D5S349, D17S250, and D2S123 are represented in Tables 3 and 4, respectively. At D5S349, 14 alleles with different allele size were identified from the normal tissue of the cancer patients. The frequency of 107 bp allele at this locus was 0.632, which was exceedingly higher than other studies in Korean population.13 The expected heterozygosity (Hexp) and PI at the D5S346 were 0.582 and 0.204, respectively. At the D17S250, there were 12 different sizes of the alleles. Besides, Hexp and PI of D17S250 were 0.742 and 0.103, respectively. At the D2S128, there were 14 different sizes alleles. Frequency of the highest and the second highest observed allele in D2S128 were 0.277 and 0.164, respectively. Furthermore, Hexp and PI of D2S128 were 0.847 and 0.044, respectively. Of the three markers, the D2S128 locus has the strongest discriminating power. The combined PI was 9.23×10-4 and combined power of exclusion was 0.99908.
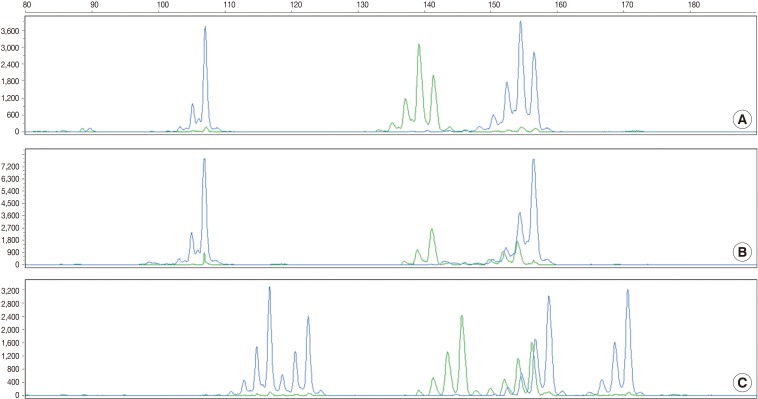
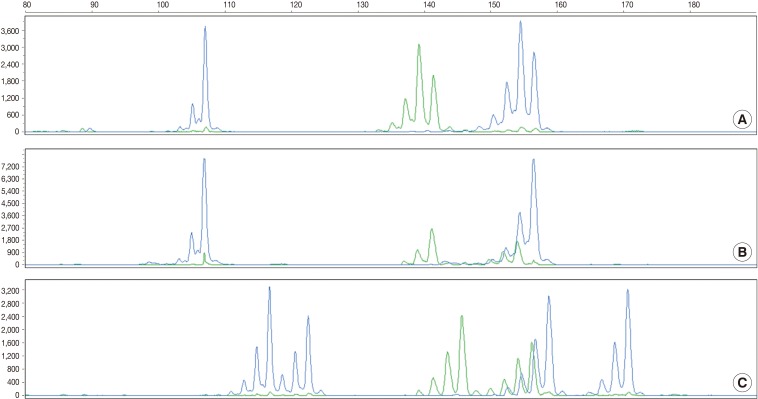
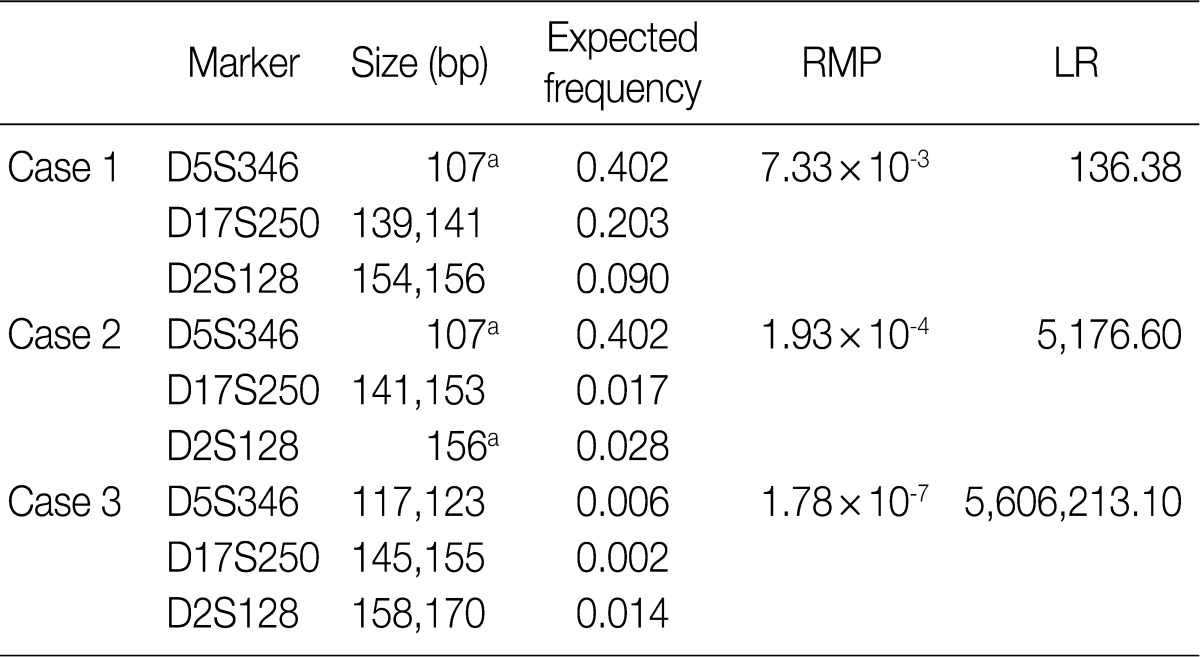
- According to the above results, we calculated the RMP and LR in all the cases. The LRs were distributed from 136.38 to 5,606,213.10. There was no case of LR<100. In addition, 56 cases (18.2%) of LR ranging from 100 to 1,000, 107 cases (42.3%) had LR ranging from 1,000 to 10,000, and 100 cases (39.5%) of LR>10,000. In general, LR of more than 10,000 was regarded as "very strong evidence to support."14 Case 1, 2, and 3 had the lowest LR, the LR of 50 percentile, and the highest LR, respectively (Table 5, Fig. 2).
- Example of SRAE
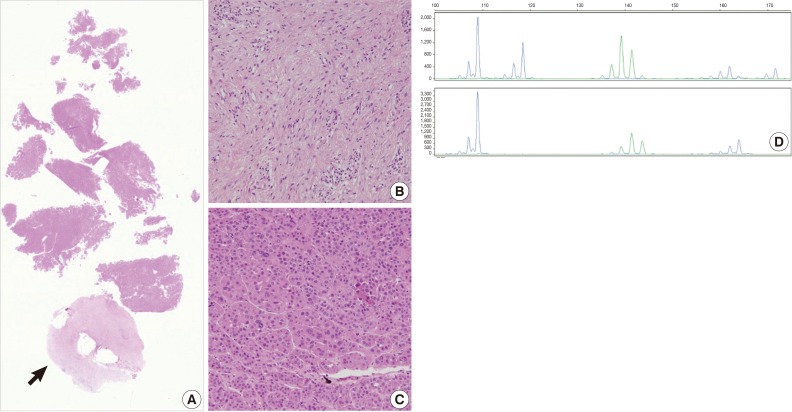
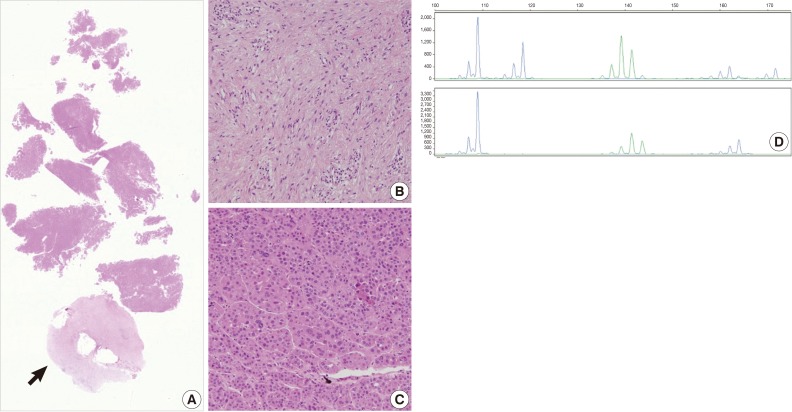
- The current case is a case suspected of specimen-mix up. The patient was operated for a slowly growing mass at the right parietal lobe of the cerebrum. Five paraffin blocks were made after gross examination, followed by preparation of hematoxylin and eosin slide. Of the 5 slides, one showed quite different histologic features from the others (Fig. 3A). It showed a fibrotic change with infiltrations of the acute and chronic inflammatory cells (Fig. 3B and arrow in Fig. 3A). These histologic features were similar to those of other slides from the same patients. But most of the fragmented tissues were composed of tumor cells with trabecular and nested pattern (Fig. 3C). Tumor cells showed polygonal shape, abundant and eosinophilic cytoplasm, pleomorphic nuclei and an immunoreactivity to anti-hepatocyte antigen. These findings are suggestive of well-differentiated hepatocellular carcinoma. The patient was young and had no history of hepatitis. In addition, the patients had a normal range of alpha-feto protein serologic test. We, therefore, considered the above histopathologic findings as a contamination rather than a metastatic lesion. This was followed by MSI tests to confirm whether the patient actually had hepatocellular carcinoma. But the MSI test results showed that there were differences in the profiles of MSI marker between the different tissue from the same slide (Fig. 3D). This led to a conclusion that the hepatocellular carcinoma tissues were included due to contamination of the tissue samples from other patient.
RESULTS
- SRAEs are critical errors that might occur in surgical pathology laboratories. If there are any chances that biopsy or cytology specimens are mixed up with each other, this would lead to the resection of normal organ or loss of possibility that patients with cancer might not be treated at the appropriate time. This happens very infrequently in clinical practice, but it is a nightmare of the pathologists and it is not absolutely avoidable. In routine laboratory procedures, there is no practical way to prove or confirm the cross-contamination or shift of the the samples.
- It would therefore, be very helpful if the surgical laboratories can set up the method which can be performed in their own laboratory for the screening the personal identity. The methods should be preferably easy and cheap. In the current experimental study, the MSI test based on three dinucleotide markers was effective in screening the personal identity of the patient as compared with other commercially available kit based on three markers (GenePrint STR Systems, Promega). Because we used only three markers for MSI test, our combined PI was lower than that of commercial kits based on multiple markers. Nevertheless, there were no cases of LR of <100 and about one-third of total cases had LR of >10,000. This suggest that there is less than 1% of chance that the patients might be different if the two samples showed the same pattern of alleles. In one third cases, this chance is less than 0.01%. In cases of specimen mixed-up, the MSI test can be considered as a successful method for screening the specimen identity.
- For the personal identification, some attentions should be paid to the interpretation of allele patterns of the MSI markers. Theoretically, the same allele pattern of the two samples does not always mean that two samples were derived from the same person. If necessary, additional tests using more STR markers should be performed. In most cases, however, we did not have to test more markers to identify the samples. In making a comparison of the allele patterns, pathologic lesion should be avoided not to get ambiguous results. There are two kinds of possibilities that diseased tissue can be disguised as the samples from the different patient. One possibility is that the homozygote locus may be mistaken as heterozygous due to MSI and the other possibility is that the heterozygous locus may be mistaken for homozygous due to a loss of heterozygosity. These types of alterations frequently occur in neoplastic lesions. Not only some types carcinomas but also some dysplastic lesions such as adenomatous lesions of colorectum and dysplasia in Barrett esophagus can be associated with the above alteration.15,16
- Currently, the local southern sizing algorithm is widely used as the standard method for measuring the length of nucleotides. In the current study, however, there was a slight difference between the estimated nucleotide length and reference nucleotide length which showed gradual change in each marker. We recommend to use the calibrated length and conservative interpretation. Sometimes repeat slippage/polymerase stuttering phenomenon was useful for correction.
- In conclusion, our results showed that the MSI test are useful to screen the personal identity in case of SRAEs in which the samples are suspected to be mixed up or shifted.
DISCUSSION
- 1. Dunn EJ, Moga PJ. Patient misidentification in laboratory medicine: a qualitative analysis of 227 root cause analysis reports in the Veterans Health Administration. Arch Pathol Lab Med 2010; 134: 244-255. ArticlePubMedPDF
- 2. Wagar EA, Stankovic AK, Raab S, Nakhleh RE, Walsh MK. Specimen labeling errors: a Q-probes analysis of 147 clinical laboratories. Arch Pathol Lab Med 2008; 132: 1617-1622. ArticlePubMedPDF
- 3. Marberger M, McConnell JD, Fowler I, et al. Biopsy misidentification identified by DNA profiling in a large multicenter trial. J Clin Oncol 2011; 29: 1744-1749. ArticlePubMedPMC
- 4. Hunt JL, Swalsky P, Sasatomi E, Niehouse L, Bakker A, Finkelstein SD. A microdissection and molecular genotyping assay to confirm the identity of tissue floaters in paraffin-embedded tissue blocks. Arch Pathol Lab Med 2003; 127: 213-217. ArticlePubMedPDF
- 5. Bell CG, Wood DR, Cheong SJ, et al. Molecular confirmation of pathological specimen integrity in Australasia. Pathology 2009; 41: 280-283. ArticlePubMed
- 6. Motani-Saitoh H, Inoue H, Tanizawa T, et al. Useful DNA typing using AmpFlSTR Identifiler Kit for formaldehyde-fixed paraffin-embedded (FFPE) tissues in early gastric cancer patient with lymph node metastasis. Histol Histopathol 2009; 24: 1139-1145. PubMed
- 7. Hunt JL. Identifying cross contaminants and specimen mix-ups in surgical pathology. Adv Anat Pathol 2008; 15: 211-217. ArticlePubMed
- 8. Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998; 58: 5248-5257. PubMed
- 9. Choi JS, Kim MA, Lee HE, Lee HS, Kim WH. Mucinous gastric carcinomas: clinicopathologic and molecular analyses. Cancer 2009; 115: 3581-3590. ArticlePubMed
- 10. Elder JK, Southern EM. Measurement of DNA length by gel electrophoresis II: Comparison of methods for relating mobility to fragment length. Anal Biochem 1983; 128: 227-231. ArticlePubMed
- 11. R: A language and environment for statistical computing. ISBN 3-900051-07-0 [Internet]. R Development Core Team. 2011; cited 2011 Mar 1. Vienna: R Foundation for Statistical Computing, Available from: http://www.R-project.org/.
- 12. Committee on DNA Forensic Science: An Update, National Research Council. The evaluation of forensic DNA evidence. 1996; Washington, DC: National Academies Press.
- 13. Kim YL, Hwang JY, Kim YJ, et al. Allele frequencies of 15 STR loci using AmpF/STR Identifiler kit in a Korean population. Forensic Sci Int 2003; 136: 92-95. ArticlePubMed
- 14. Evett IW, Jackson G, Lambert JA, McCrossan S. The impact of the principles of evidence interpretation on the structure and content of statements. Sci Justice 2000; 40: 233-239. ArticlePubMed
- 15. Yanagi M, Keller G, Mueller J, et al. Comparison of loss of heterozygosity and microsatellite instability in adenocarcinomas of the distal esophagus and proximal stomach. Virchows Arch 2000; 437: 605-610. ArticlePubMedPDF
- 16. Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer 2009; 9: 489-499. ArticlePubMedPDF
REFERENCES
aExpected heterozygosity; bPolymorphism information content (the probability that a given offspring of a parent carrying a rare allele at a locus will allow deduction of the parental genotype at the locus); cPower of discrimination (defined as PD=1-PI); dProbability of identity (matching probability, the probability that two individuals selected at random will have an identical genotype at the tested locus).
Figure & Data
References
Citations

- Lost, mislabeled, and mishandled surgical and clinical pathology specimens: A systematic review of published literature
Heather J Carmack, Braidyn S Lazenby, Kylie J Wilson, Jamie N Bakkum-Gamez, Leslie Carranza
American Journal of Clinical Pathology.2024; 162(4): 349. CrossRef - Sensitivity and polymorphism of Bethesda panel markers in Chinese population
Yanying Zheng, Jie Chen, Xiang Zhang, Ling Xie, Yifen Zhang, Yi Sun
Bulletin du Cancer.2020; 107(11): 1091. CrossRef
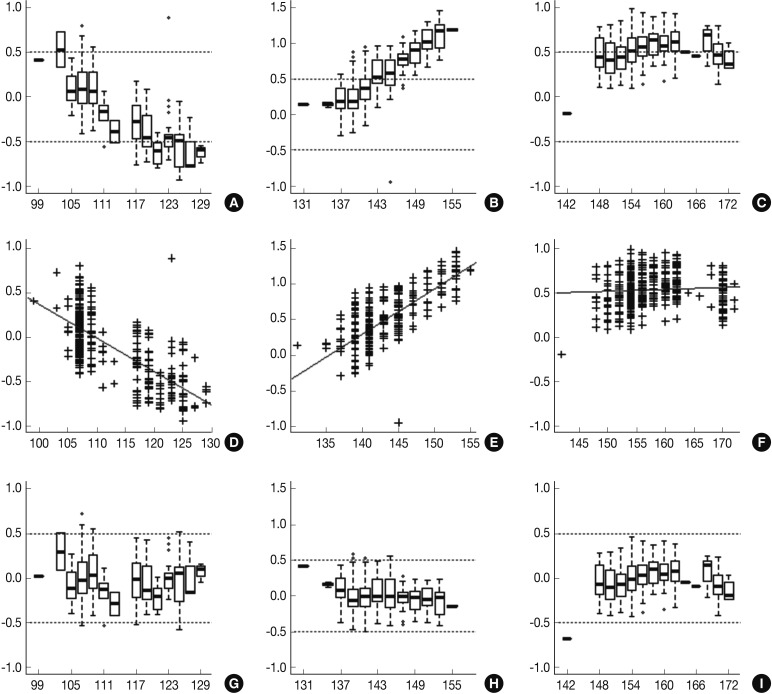


Fig. 1
Fig. 2
Fig. 3




Minimum allele frequency: 0.010 (When allele frequencies of estimated nucleotides length fall below the minimum allele frequency, then the minimum allele frequency is used instead).
aExpected heterozygosity; bPolymorphism information content (the probability that a given offspring of a parent carrying a rare allele at a locus will allow deduction of the parental genotype at the locus); cPower of discrimination (defined as PD=1-PI); dProbability of identity (matching probability, the probability that two individuals selected at random will have an identical genotype at the tested locus).
RMP, random match probability; LR, likelihood ratio. aHomozygote.

 E-submission
E-submission


 PubReader
PubReader Cite this Article
Cite this Article