Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 46(2); 2012 > Article
-
Original Article
Identifying Polymorphisms inIL-31 and Their Association with Susceptibility to Asthma - Ji-In Yu1, Weon-Cheol Han1, Ki-Jung Yun1,2, Hyung-Bae Moon1, Gyung-Jae Oh3, Soo-Cheon Chae1,2
-
Korean Journal of Pathology 2012;46(2):162-168.
DOI: https://doi.org/10.4132/KoreanJPathol.2012.46.2.162
Published online: April 25, 2012
1Department of Pathology, Wonkwang University School of Medicine, Iksan, Korea.
2Digestive Disease Research Institute, Wonkwang University, Iksan, Korea.
3Department of Preventive Medicine, Wonkwang University School of Medicine, Iksan, Korea.
- Corresponding Author: Soo-Cheon Chae, Ph.D. Department of Pathology, Wonkwang University School of Medicine, 895 Muwang-ro, Iksan 570-749, Korea. Tel: +82-63-850-6793, Fax: +82-63-852-2110, chaesc@wku.ac.kr
© 2012 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- Interleukin 31 (IL-31) is a T helper type 2 effector cytokine that plays an important role in the pathogenesis of atopic and allergic diseases. IL-31 may be involved in promoting allergic inflammation and in inducing airway epithelial responses such as allergic asthma.
-
Methods
- Single-base extension analysis was used to detect the genotypes of IL-31 single nucleotide polymorphisms (SNPs), and we compared the genotype and allele frequencies of the IL-31 SNPs between patients with asthma and healthy controls.
-
Results
- There were no significant differences in the genotype and allele frequencies of the IL-31 SNPs between patients with asthma and healthy controls. Furthermore we compared the genotype and allele frequencies of IL-31 SNPs between patients with atopic asthma, those with non-atopic asthma and healthy controls. This showed that the SNPs were not associated with the susceptibility to atopic asthma. There were no significant differences in the haplotype frequencies of IL-31 SNPs between patients with asthma and healthy controls. In patients with asthma, the IL-31 SNPs were significantly correlated with total serum levels of IgE (p=0.035).
-
Conclusions
- Our results indicate that, the IL-31 SNPs may be associated with IgE production in patients with asthma.
- Patients and DNA samples
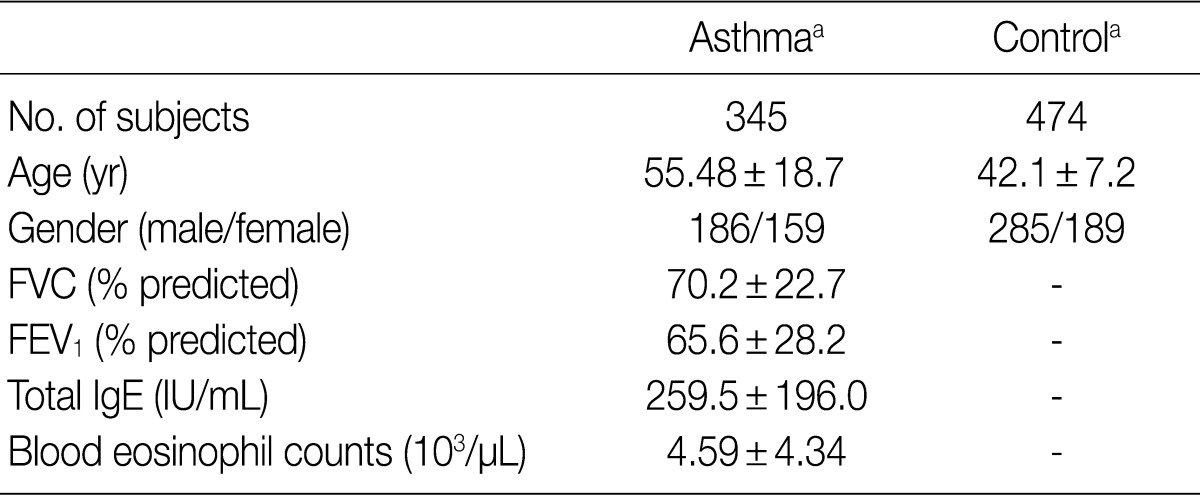
- The DNA samples used in the current study were provided by the Biobank of Wonkwang University Hospital, a member of the National Biobank of Korea; this Biobank is supported by the Ministry of Health and Welfare Affairs. The current study was approved by the Institutional Review Board (IRB) of our medical institution. All the subjects submitted a written informed consent. We obtained the genomic DNA samples from 345 patients with asthma and 474 healthy controls. The clinical parameters of the study subjects are summarized in Table 1. Genomic DNA was extracted from peripheral blood leukocytes by using a standard phenol-chloroform method or by using a genomic DNA extraction kit (iNtRON Biotechnology, Seoul, Korea) according to the manufacturer's instructions. Patients were diagnosed with asthma according to the criteria of the American Thoracic Society.15 In patients with asthma, the blood eosinophil counts and total serum IgE levels were measured using a Coulter® Gen.S™ Hematology Analyzer (Beckman, Hialeh, FL, USA) and a Roche COBAS-CORE II (Roche Diagnostics, Basal, Switzerland), respectively. All the subjects who were enrolled in the current study between January 2003 and December 2005 were Korean people living in the same area.
- Polymerase chain reaction (PCR) and sequencing analysis
- The entire coding regions of IL-31, including the 2.0 kb promoter regions, were partially amplified using two primer pairs (Table 2). Predenaturation treatment of template DNA was performed in a PCR Thermal Cycler DICE Gradient (TaKaRa, Shiga, Japan) at 95℃ for 7 minutes, which was followed by 29 cycles of denaturation at 95℃ for 10 seconds, annealing at 65℃ for 30 seconds and extension at 72℃ for 2.5 minutes. The final extension was completed at 72℃ for 10 minutes. After purification using a PCR purification kit (Millipore, Billerica, MA, USA), the PCR products were used as template DNA for sequencing analysis using the ABI Prism BigDye Terminator cycle sequencing system (PE Applied Biosystems, Corona, CA, USA) on the ABI 3100 automatic sequencer (PE Applied Biosystems). We used the same primers for PCR, and eleven additional primers were employed for the sequencing of the IL-31 gene (Table 2). Sequence analysis was performed to detect the IL-31 SNPs; the sequence of human chromosome 12 BAC RP11-512M8 was used as the reference sequence.
- Genotype analysis
- The single-base extension (SBE) method was used for the genetic analysis of g.-1550T>C, g.-1066G>A, g.586C>A, and g.1449C>G in the IL-31 gene. The PCR was performed using a 50 ng of each genomic DNA and Taq DNA polymerase (EF Taq, Solgent, Daejeon, Korea) and 0.5 µM of each primer under the following conditions: 30 cycles of denaturation at 98℃ for 10 seconds, annealing at 55℃ for 10 seconds, and extension at 72℃ for 30 seconds. The final extension was completed at 72℃ for 10 minutes in a thermocycler (PE Applied Biosystems). The PCR products were purified using a PCR purification kit (Millipore, Bedford, CA, USA) and then used as the template DNA for the SBE primers (Table 2). The SBE reaction mix was prepared according to a previously described method.16 The primer extension reaction was performed according to a previously described method.17
- Statistical analysis
- A case-control association analysis was used to compare the findings between patients with asthma and healthy controls. χ2-test was performed to estimate the Hardy-Weinberg equilibrium (HWE). A pair-wise comparison of the biallelic loci was employed to analyzed the linkage disequilibrium (LD). The haplotype frequencies for multiple loci of IL-31 were estimated with the expectation-maximization algorithm by using SNPAlyze software (DYNACOM, Yokohama, Japan). Logistic regression analysis (ver. 11.5, SPSS Inc., Chicago, IL, USA) was performed to calculate the odds ratios (with the 95% confidence intervals). Analysis of variation (ANOVA) was performed to determine the IgE levels and the peripheral blood eosinophils counts for each genotype of individual patients with asthma. A p<0.05 was considered statistically significant.
MATERIALS AND METHODS
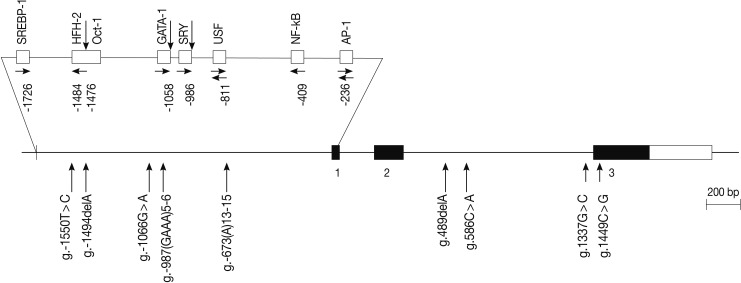
- The human IL-31 gene is located on chromosome 12q24.31, and it consists of three exons. We scanned the genomic DNA samples isolated from 24 unrelated patients with asthma and 24 healthy controls by using a direct sequencing method to determine the possible variation sites in the coding regions and boundary intron sequences of IL-31, including ~2.0 kb promoter regions. We identified five SNPs and four variation sites, i.e., g.-1550T>C (rs7312610), g.-1494delA (novel), g.-1066G>A (rs11608363), g.-987(GAAA)5-6 (novel), and g.-673(A)13-15 (novel) in the promoter region; g.489delA (novel), g.586C>A (novel), and g.1337G>C (rs7977932) in intron 2; and g.1449C> G (rs7974857) in exon 3 (Fig. 1). The g.1449C>G polymorphisms located in the coding region were synonymous SNPs (Gly61Gly) in the IL-31 gene. We calculated the LD coefficients (|D'|) between all the SNP pairs, and determined the absolute LD (|D'|=1 and r2=1) between g.-1550T>C and g.1449C>G (data not shown). Of the identified polymorphisms, three SNPs (g.-1066G>A, g.586C>A, and g.1449C>G) were selected for large-sample genotyping on the basis of their locations and LD coefficients.
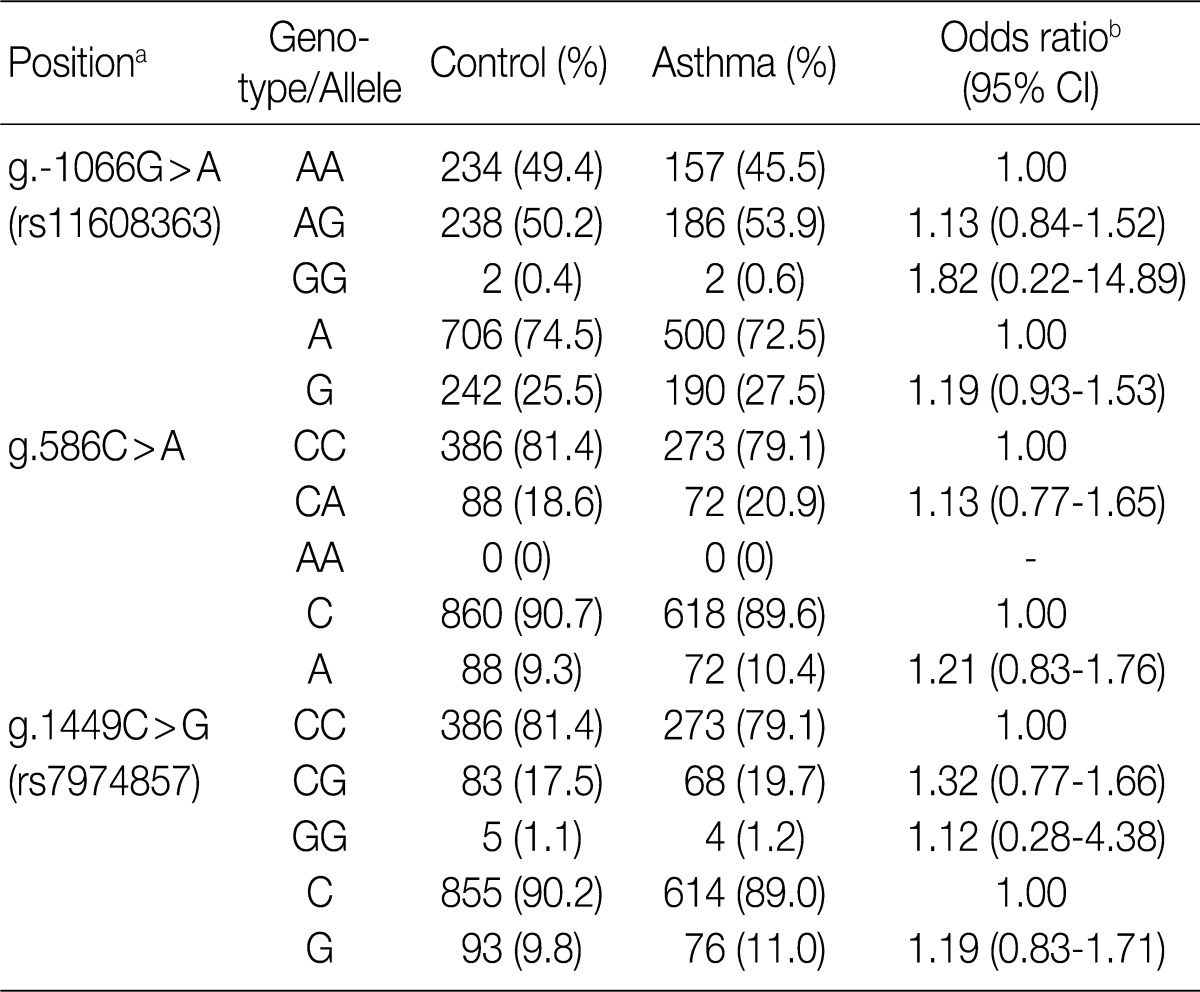
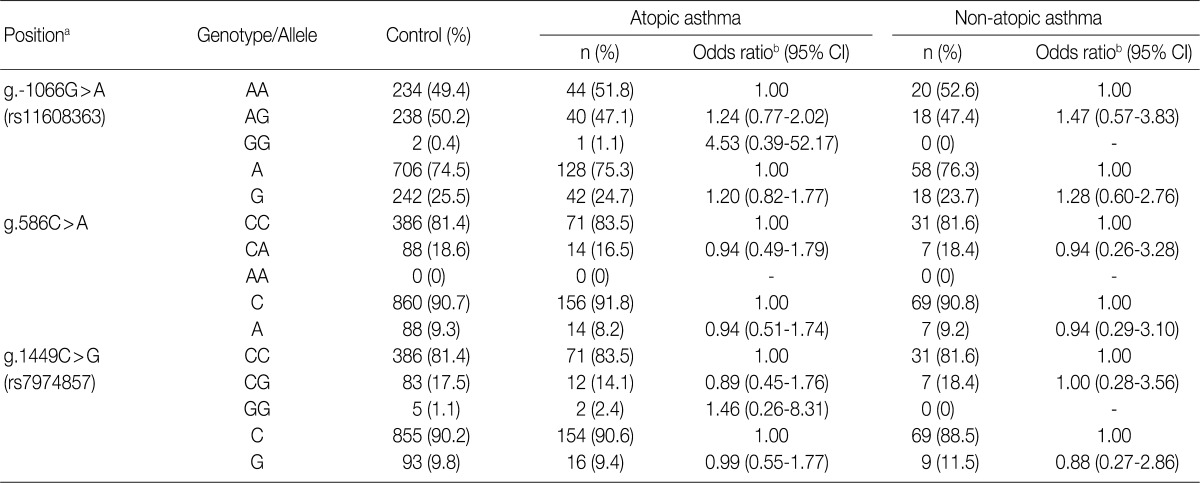
- To determine whether the IL-31 SNPs are associated with susceptibility to asthma, we analyzed their genotypes using the SBE method. We compared the genotypes and allele frequencies between the patients with asthma and the healthy controls. All the genotype frequencies in both the healthy controls and the patients with asthma were consistent with HWE, except for g.-1066G>A (data not shown). There were no significant differences in the genotype and allele frequencies of g.-1066G>A, g.586C>A, and g.1449C>G between patients with asthma and healthy controls (Table 3). We further analyzed the genotype and allele frequencies in patients with atopic asthma, those with non-atopic asthma, and healthy controls (Table 3). This showed that there were also no significant differences in the genotype and allele frequencies of g.-1066G>A, g.586C>A, and g.1449C>G between patients with atopic asthma, those with non-atopic asthma and healthy controls (Table 4). These results suggest that the IL-31 SNPs might not be associated with the susceptibility to asthma.
- We further investigated whether the IL-31 SNPs are also associated with the total serum IgE levels, peripheral blood eosinophil counts and FVC and FEV1 values in patients with asthma. In patients with asthma, the IL-31 SNPs had no significant correlation with the peripheral blood eosinophil counts and the FVC and FEV1 values (Table 5). But, the g.1449C>G (also g.-1550T>C) of IL-31 was significantly correlated with total serum IgE levels (p=0.035) (Table 5). These results indicate that the IL-31 SNPs may influence IgE production in patients with asthma.
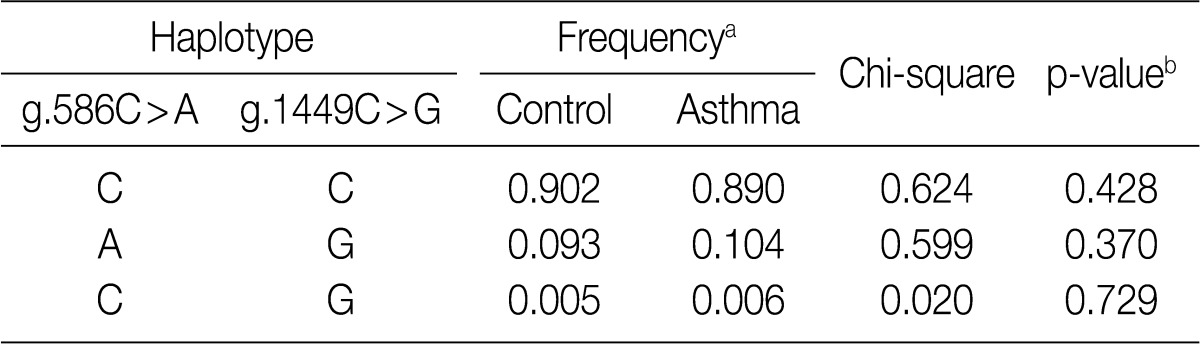
- Finally, we compared the haplotype frequencies of the g.586C>A and g.1449C>G SNPs of IL-31 in both healthy controls and patients with asthma (Table 6). There were no significant differences in the major and minor haplotype frequencies between the patients with asthma and healthy controls. These results suggest that the haplotypes of the IL-31 polymorphisms are not correlated with the susceptibility to asthma.
RESULTS
- Asthma is a chronic allergic inflammatory disease of the airway, and it is characterized by bronchial infiltration of eosinophils, elevated levels of both IgE and Th2 cytokines, reversible-airflow obstruction, mucus hypersecretion, and bronchial hyperreactivity.3 In the stage of pathogenesis, the activated Th cells differentiate into two different types of cells, both phenotypically and functionally, and these include Th1 and Th2 cells.18,19 Th1 cells produce cytokines such as interferon-γ, IL-12 and cytotoxic factor lymphotoxin. These cells are commonly associated with cell-mediated immune responses against intracellular pathogens and induction of organ-specific autoimmune diseases.19,20 In contrast, Th2 cells produce cytokines such as IL-4, IL-5, and IL-10; these cytokines are associated with atopic and allergic diseases such as asthma. In our previous studies on Korean population, we found that the SNPs or genetic variations in Tim-1 and IL-27 are associated with the susceptibility to asthma.21,22
- IL-31 is believed to play an important role in promoting allergic inflammation and inducing airway epithelial response such as allergic asthma.11 In the current study, we evaluated the associations between IL-31 polymorphisms and the susceptibility to asthma. We not only identified five SNPs, including a novel SNP and four novel variation sites in the IL-31 gene, but also analyzed the genotypes of the g.-1066G>A, g.586C>A, and g.1449C>G SNPs in patients with asthma and healthy controls. In addition, there were no significant differences in the genotype and allele frequencies of the IL-31 SNPs between patients with asthma and healthy controls (Table 3). We also compared the genotype and allele frequencies between patients with atopic asthma, those with non-atopic asthma and healthy controls. This showed that the genotype and allele frequencies of the IL-31 SNPs were not associated with atopic asthma (Table 4). These results suggest that the IL-31 SNPs may not be associated with the susceptibility to asthma.
- A large number of eosinophils are accumulated in the lungs of patients with asthma, and they are essential for phagocytosis as well as the allergic and inflammatory reactions of asthma. At least dozens of polymorphic genes have been reported to regulate asthma by controlling the inflammatory response and the serum levels of IgE, cytokines, and chemokines.23 We and other research groups have previously shown that the SNPs of eotaxin gene family are associated with total serum levels of IgE in patients with asthma.24,25 Shin et al. suggested that IL-18 polymorphisms are associated with specific levels of IgE to mite allergens in patients with asthma.26 Our results revealed that the IL-31 SNPs in patients with asthma are closely associated with total serum level of IgE, but not with the peripheral blood eosinophil counts and the FVC and FEV1 values (Table 5). These results indicate that IL-31 SNPs might be related to total serum levels of IgE in asthmatic response and the activation of mast cells. The genotype of IL-31 SNPs might have a relationship with the occurrence of asthmatic symptoms or their severity. This is not notable not only because IL-31 is mainly produced by activated Th2 cells but also because the production of IgE is initiated by Th2 cells.
- It has recently been reported that nonatopic eczema is strongly associated with the following mutations in a common risk haplotype GAA of IL-31: IL-31-2057G>A (rs6489188), IL-31-1066G>A (rs11608363), and IL-31IVS2+12A>G of IL-31. Besides, the degree of IL-31 expression was significantly higher in carriers of the risk haplotype as compared with non-carriers.14 Further, a single SNP in the promoter region may affect the regulation of IL-31 expression.14 Our results showed that the g.-1550T>C (rs7312610) and g.1449C>G (rs7974857) of the IL-31 were significantly associated with total serum levels of IgE (Table 5). This result indicates that IL-31 polymorphism may be associated with IgE production.
- In conclusion, our results suggest that IL-31 may be a candidate gene that is associated with the production of IgE in asthma. But we could not completely rule out the possibility that multiple genetic alterations might also lead to the aggravation of asthma.
DISCUSSION
Acknowledgments
Acknowledgments
- 1. Cookson W. Genetics and genomics of asthma and allergic diseases. Immunol Rev 2002; 190: 195-206. ArticlePubMedPDF
- 2. Fahy JV, Corry DB, Boushey HA. Airway inflammation and remodeling in asthma. Curr Opin Pulm Med 2000; 6: 15-20. ArticlePubMed
- 3. Chiappara G, Gagliardo R, Siena A, et al. Airway remodelling in the pathogenesis of asthma. Curr Opin Allergy Clin Immunol 2001; 1: 85-93. ArticlePubMed
- 4. Koyama S, Sato E, Tsukadaira A, et al. Vascular endothelial growth factor mRNA and protein expression in airway epithelial cell lines in vitro. Eur Respir J 2002; 20: 1449-1456. ArticlePubMed
- 5. Coyle AJ, Wagner K, Bertrand C, Tsuyuki S, Bews J, Heusser C. Central role of immunoglobulin (Ig) E in the induction of lung eosinophil infiltration and T helper 2 cell cytokine production: inhibition by a non-anaphylactogenic anti-IgE antibody. J Exp Med 1996; 183: 1303-1310. ArticlePubMedPMCPDF
- 6. Daser A, Meissner N, Herz U, Renz H. Role and modulation of T-cell cytokines in allergy. Curr Opin Immunol 1995; 7: 762-770. ArticlePubMed
- 7. O'Garra A. Cytokines induce the development of functionally heterogeneous T helper cell subsets. Immunity 1998; 8: 275-283. ArticlePubMed
- 8. Humbles AA, Conroy DM, Marleau S, et al. Kinetics of eotaxin generation and its relationship to eosinophil accumulation in allergic airways disease: analysis in a guinea pig model in vivo. J Exp Med 1997; 186: 601-612. ArticlePubMedPMCPDF
- 9. Dillon SR, Sprecher C, Hammond A, et al. Interleukin 31, a cytokine produced by activated T cells, induces dermatitis in mice. Nat Immunol 2004; 5: 752-760. ArticlePubMedPDF
- 10. Sonkoly E, Muller A, Lauerma AI, et al. IL-31: a new link between T cells and pruritus in atopic skin inflammation. J Allergy Clin Immunol 2006; 117: 411-417. ArticlePubMed
- 11. Chattopadhyay S, Tracy E, Liang P, Robledo O, Rose-John S, Baumann H. Interleukin-31 and oncostatin-M mediate distinct signaling reactions and response patterns in lung epithelial cells. J Biol Chem 2007; 282: 3014-3026. ArticlePubMed
- 12. Ip WK, Wong CK, Li ML, Li PW, Cheung PF, Lam CW. Interleukin-31 induces cytokine and chemokine production from human bronchial epithelial cells through activation of mitogen-activated protein kinase signalling pathways: implications for the allergic response. Immunology 2007; 122: 532-541. ArticlePubMedPMC
- 13. Lei Z, Liu G, Huang Q, et al. SCF and IL-31 rather than IL-17 and BAFF are potential indicators in patients with allergic asthma. Allergy 2008; 63: 327-332. ArticlePubMed
- 14. Schulz F, Marenholz I, Folster-Holst R, et al. A common haplotype of the IL-31 gene influencing gene expression is associated with nonatopic eczema. J Allergy Clin Immunol 2007; 120: 1097-1102. ArticlePubMed
- 15. Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease (COPD) and asthma. This official statement of the American Thoracic Society was adopted by the ATS Board of Directors, November 1986. Am Rev Respir Dis 1987; 136: 225-244. ArticlePubMed
- 16. Li CS, Zhang Q, Lim MK, et al. Association of FOXJ1 polymorphisms with systemic lupus erythematosus and rheumatoid arthritis in Korean population. Exp Mol Med 2007; 39: 805-811. ArticlePubMedPDF
- 17. Chae SC, Shim SC, Chung HT. Association of TBX21 polymorphisms in a Korean population with rheumatoid arthritis. Exp Mol Med 2009; 41: 33-41. ArticlePubMedPMC
- 18. Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol 1989; 7: 145-173. ArticlePubMed
- 19. Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature 1996; 383: 787-793. ArticlePubMedPDF
- 20. Kuchroo VK, Das MP, Brown JA, et al. B7-1 and B7-2 costimulatory molecules activate differentially the Th1/Th2 developmental pathways: application to autoimmune disease therapy. Cell 1995; 80: 707-718. ArticlePubMed
- 21. Chae SC, Song JH, Heo JC, Lee YC, Kim JW, Chung HT. Molecular variations in the promoter and coding regions of human Tim-1 gene and their association in Koreans with asthma. Hum Immunol 2003; 64: 1177-1182. ArticlePubMed
- 22. Chae SC, Li CS, Kim KM, et al. Identification of polymorphisms in human interleukin-27 and their association with asthma in a Korean population. J Hum Genet 2007; 52: 355-361. ArticlePubMedPDF
- 23. Cookson W. The alliance of genes and environment in asthma and allergy. Nature 1999; 402(6760 Suppl):B5-B11. ArticlePubMedPDF
- 24. Chae SC, Lee YC, Park YR, et al. Analysis of the polymorphisms in eotaxin gene family and their association with asthma, IgE, and eosinophil. Biochem Biophys Res Commun 2004; 320: 131-137. ArticlePubMed
- 25. Shin HD, Kim LH, Park BL, et al. Association of Eotaxin gene family with asthma and serum total IgE. Hum Mol Genet 2003; 12: 1279-1285. ArticlePubMed
- 26. Shin HD, Kim LH, Park BL, et al. Association of interleukin 18 (IL18) polymorphisms with specific IgE levels to mite allergens among asthmatic patients. Allergy 2005; 60: 900-906. ArticlePubMed
REFERENCES

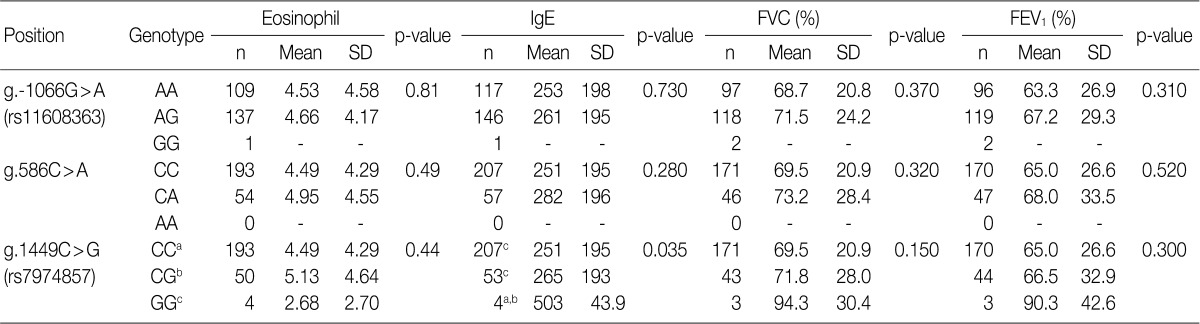
Data are analyzed by ANOVA. Significant differences (p<0.05) between two groups found by Bonferroni multiple comparisons are indicated by values which have the same letter (a-c). The position is calculated from the translation start site.
FVC, forced vital capacity; FEV1, forced expiratory volume in 1 sec; SD, standard deviation.
Figure & Data
References
Citations

- IL-31: State of the Art for an Inflammation-Oriented Interleukin
Francesco Borgia, Paolo Custurone, Federica Li Pomi, Raffaele Cordiano, Clara Alessandrello, Sebastiano Gangemi
International Journal of Molecular Sciences.2022; 23(12): 6507. CrossRef - Interleukin-31 and soluble CD40L: new candidate serum biomarkers that predict therapeutic response in multiple sclerosis
Isabelle Pastor Bandeira, André Eduardo de Almeida Franzoi, Giulia Murillo Wollmann, Washigton Luiz Gomes de Medeiros Junior, Wesley Nogueira Brandão, Jean Pierre Schatzmann Peron, Jefferson Becker, Osvaldo José Moreira Nascimento, Marcus Vinícius Magno G
Neurological Sciences.2022; 43(11): 6271. CrossRef - Interleukin‐31: The “itchy” cytokine in inflammation and therapy
Angeliki Datsi, Martin Steinhoff, Fareed Ahmad, Majid Alam, Joerg Buddenkotte
Allergy.2021; 76(10): 2982. CrossRef - Infection-Associated Mechanisms of Neuro-Inflammation and Neuro-Immune Crosstalk in Chronic Respiratory Diseases
Belinda Camp, Sabine Stegemann-Koniszewski, Jens Schreiber
International Journal of Molecular Sciences.2021; 22(11): 5699. CrossRef - Livestock farm particulate matter enhances airway inflammation in mice with or without allergic airway disease
Dingyu Liu, James G. Wagner, Jack R. Harkema, Miriam E. Gerlofs-Nijland, Elena Pinelli, Gert Folkerts, Rob J. Vandebriel, Flemming R. Cassee
World Allergy Organization Journal.2020; 13(4): 100114. CrossRef - IL-31: A new key player in dermatology and beyond
Işın Sinem Bağci, Thomas Ruzicka
Journal of Allergy and Clinical Immunology.2018; 141(3): 858. CrossRef - The Role of Interleukin-31 Polymorphisms in Non-Small Cell Lung Cancer Genetic Susceptibility and Clinical Outcome
Yongfeng Yang, Li Li, Fei Chen, Li Zhang, Hong Bu
Genetic Testing and Molecular Biomarkers.2018; 22(5): 314. CrossRef - Associations betweenInterleukin-31Gene Polymorphisms and Dilated Cardiomyopathy in a Chinese Population
Huizi Song, Ying Peng, Bin Zhou, Nan Chen, Xiaochuan Xie, Qingyu Dou, Yue Zhong, Li Rao
Disease Markers.2017; 2017: 1. CrossRef - The association of interleukin-31 polymorphisms with interleukin-31 serum levels and risk of systemic lupus erythematosus
Hua-Tuo Huang, Jian-Ming Chen, Jing Guo, Yan Lan, Ye-Sheng Wei
Rheumatology International.2016; 36(6): 799. CrossRef - Stem Cell Factor and Interleukin-31 Expression: Association with IgE among Egyptian Patients with Atopic and Nonatopic Bronchial Asthma
M. Moaaz, S. Abo El-Nazar, M. Abd El-Rahman, E. Soliman
Immunological Investigations.2016; 45(2): 87. CrossRef - Interleukin-31 expression and relation to disease severity in human asthma
Tianwen Lai, Dong Wu, Wen Li, Min Chen, Zhennan Yi, Dan Huang, Zhiliang Jing, Yingying Lü, Quanchao Lv, Dongming Li, Bin Wu
Scientific Reports.2016;[Epub] CrossRef - Elevated TGF-β1/IL-31 Pathway Is Associated with the Disease Severity of Hepatitis B Virus–Related Liver Cirrhosis
Desong Ming, Xueping Yu, Ruyi Guo, Yong Deng, Julan Li, Chengzu Lin, Milong Su, Zhenzhong Lin, Zhijun Su
Viral Immunology.2015; 28(4): 209. CrossRef - Interleukin-31 promotes helper T cell type-2 inflammation in children with allergic rhinitis
Wenlong Liu, Renzhong Luo, Yanqiu Chen, Changzhi Sun, Jie Wang, Lifeng Zhou, Yan Li, Li Deng
Pediatric Research.2015; 77(1): 20. CrossRef - The Transforming Growth Factor β1/Interleukin-31 Pathway Is Upregulated in Patients with Hepatitis B Virus-Related Acute-on-Chronic Liver Failure and Is Associated with Disease Severity and Survival
Xueping Yu, Ruyi Guo, Desong Ming, Yong Deng, Milong Su, Chengzu Lin, Julan Li, Zhenzhong Lin, Zhijun Su, R. L. Hodinka
Clinical and Vaccine Immunology.2015; 22(5): 484. CrossRef - NFAT1 and JunB Cooperatively Regulate IL-31 Gene Expression in CD4+ T Cells in Health and Disease
Ji Sun Hwang, Gi-Cheon Kim, EunBee Park, Jung-Eun Kim, Chang-Suk Chae, Won Hwang, Changhon Lee, Sung-Min Hwang, Hui Sun Wang, Chang-Duk Jun, Dipayan Rudra, Sin-Hyeog Im
The Journal of Immunology.2015; 194(4): 1963. CrossRef - Interleukin-31: A Novel Diagnostic Marker of Allergic Diseases
Anja Rabenhorst, Karin Hartmann
Current Allergy and Asthma Reports.2014;[Epub] CrossRef - Role of IL-31 in regulation of Th2 cytokine levels in patients with nasal polyps
Hong Ouyang, Jie Cheng, Yajun Zheng, Jingdong Du
European Archives of Oto-Rhino-Laryngology.2014; 271(10): 2703. CrossRef - Polymorphisms of interleukin-31 are associated with anti-CCP levels in females with rheumatoid arthritis
JI-IN YU, YOUNG-RAN PARK, SHIN-SEOK LEE, SOO-CHEON CHAE
Journal of Genetics.2014; 93(3): 813. CrossRef - IL-31 Associated with Coronary Artery Lesion Formation in Kawasaki Disease
Wan-Ning Tseng, Mao-Hung Lo, Mindy Ming-Huey Guo, Kai-Sheng Hsieh, Wei-Chiao Chang, Ho-Chang Kuo, Chien-Sheng Chen
PLoS ONE.2014; 9(8): e105195. CrossRef

Fig. 1






FVC, forced vital capacity; FEV1, forced expiratory volume in 1 sec. aValues are presented as means±standard deviation.
PCR, polymerase chain reaction;
aCalculated from the translation start site; bLogistic regression analysis was used for calculating odds ratio (95% confidence interval [CI]), the results were adjusted for age.
aCalculated from the translation start site; bLogistic regression analysis was used for calculating odds ratio (95% confidence interval [CI]), the results were adjusted for age.
Data are analyzed by ANOVA. Significant differences (p<0.05) between two groups found by Bonferroni multiple comparisons are indicated by values which have the same letter (a-c). The position is calculated from the translation start site. FVC, forced vital capacity; FEV1, forced expiratory volume in 1 sec; SD, standard deviation.
aValues are constructed by expectation-maximization algorithm with genotyped SNPs; bValues are analyzed by permutation test.

 E-submission
E-submission

 PubReader
PubReader Cite this Article
Cite this Article