Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 46(2); 2012 > Article
-
Original Article
Identifying Polymorphisms inIL-31 and Their Association with Susceptibility to Asthma - Ji-In Yu1, Weon-Cheol Han1, Ki-Jung Yun1,2, Hyung-Bae Moon1, Gyung-Jae Oh3, Soo-Cheon Chae1,2
-
Korean Journal of Pathology 2012;46(2):162-168.
DOI: https://doi.org/10.4132/KoreanJPathol.2012.46.2.162
Published online: April 25, 2012
1Department of Pathology, Wonkwang University School of Medicine, Iksan, Korea.
2Digestive Disease Research Institute, Wonkwang University, Iksan, Korea.
3Department of Preventive Medicine, Wonkwang University School of Medicine, Iksan, Korea.
- Corresponding Author: Soo-Cheon Chae, Ph.D. Department of Pathology, Wonkwang University School of Medicine, 895 Muwang-ro, Iksan 570-749, Korea. Tel: +82-63-850-6793, Fax: +82-63-852-2110, chaesc@wku.ac.kr
© 2012 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Figure & Data
References
Citations

- IL-31: State of the Art for an Inflammation-Oriented Interleukin
Francesco Borgia, Paolo Custurone, Federica Li Pomi, Raffaele Cordiano, Clara Alessandrello, Sebastiano Gangemi
International Journal of Molecular Sciences.2022; 23(12): 6507. CrossRef - Interleukin-31 and soluble CD40L: new candidate serum biomarkers that predict therapeutic response in multiple sclerosis
Isabelle Pastor Bandeira, André Eduardo de Almeida Franzoi, Giulia Murillo Wollmann, Washigton Luiz Gomes de Medeiros Junior, Wesley Nogueira Brandão, Jean Pierre Schatzmann Peron, Jefferson Becker, Osvaldo José Moreira Nascimento, Marcus Vinícius Magno G
Neurological Sciences.2022; 43(11): 6271. CrossRef - Interleukin‐31: The “itchy” cytokine in inflammation and therapy
Angeliki Datsi, Martin Steinhoff, Fareed Ahmad, Majid Alam, Joerg Buddenkotte
Allergy.2021; 76(10): 2982. CrossRef - Infection-Associated Mechanisms of Neuro-Inflammation and Neuro-Immune Crosstalk in Chronic Respiratory Diseases
Belinda Camp, Sabine Stegemann-Koniszewski, Jens Schreiber
International Journal of Molecular Sciences.2021; 22(11): 5699. CrossRef - Livestock farm particulate matter enhances airway inflammation in mice with or without allergic airway disease
Dingyu Liu, James G. Wagner, Jack R. Harkema, Miriam E. Gerlofs-Nijland, Elena Pinelli, Gert Folkerts, Rob J. Vandebriel, Flemming R. Cassee
World Allergy Organization Journal.2020; 13(4): 100114. CrossRef - IL-31: A new key player in dermatology and beyond
Işın Sinem Bağci, Thomas Ruzicka
Journal of Allergy and Clinical Immunology.2018; 141(3): 858. CrossRef - The Role of Interleukin-31 Polymorphisms in Non-Small Cell Lung Cancer Genetic Susceptibility and Clinical Outcome
Yongfeng Yang, Li Li, Fei Chen, Li Zhang, Hong Bu
Genetic Testing and Molecular Biomarkers.2018; 22(5): 314. CrossRef - Associations betweenInterleukin-31Gene Polymorphisms and Dilated Cardiomyopathy in a Chinese Population
Huizi Song, Ying Peng, Bin Zhou, Nan Chen, Xiaochuan Xie, Qingyu Dou, Yue Zhong, Li Rao
Disease Markers.2017; 2017: 1. CrossRef - The association of interleukin-31 polymorphisms with interleukin-31 serum levels and risk of systemic lupus erythematosus
Hua-Tuo Huang, Jian-Ming Chen, Jing Guo, Yan Lan, Ye-Sheng Wei
Rheumatology International.2016; 36(6): 799. CrossRef - Stem Cell Factor and Interleukin-31 Expression: Association with IgE among Egyptian Patients with Atopic and Nonatopic Bronchial Asthma
M. Moaaz, S. Abo El-Nazar, M. Abd El-Rahman, E. Soliman
Immunological Investigations.2016; 45(2): 87. CrossRef - Interleukin-31 expression and relation to disease severity in human asthma
Tianwen Lai, Dong Wu, Wen Li, Min Chen, Zhennan Yi, Dan Huang, Zhiliang Jing, Yingying Lü, Quanchao Lv, Dongming Li, Bin Wu
Scientific Reports.2016;[Epub] CrossRef - Elevated TGF-β1/IL-31 Pathway Is Associated with the Disease Severity of Hepatitis B Virus–Related Liver Cirrhosis
Desong Ming, Xueping Yu, Ruyi Guo, Yong Deng, Julan Li, Chengzu Lin, Milong Su, Zhenzhong Lin, Zhijun Su
Viral Immunology.2015; 28(4): 209. CrossRef - Interleukin-31 promotes helper T cell type-2 inflammation in children with allergic rhinitis
Wenlong Liu, Renzhong Luo, Yanqiu Chen, Changzhi Sun, Jie Wang, Lifeng Zhou, Yan Li, Li Deng
Pediatric Research.2015; 77(1): 20. CrossRef - The Transforming Growth Factor β1/Interleukin-31 Pathway Is Upregulated in Patients with Hepatitis B Virus-Related Acute-on-Chronic Liver Failure and Is Associated with Disease Severity and Survival
Xueping Yu, Ruyi Guo, Desong Ming, Yong Deng, Milong Su, Chengzu Lin, Julan Li, Zhenzhong Lin, Zhijun Su, R. L. Hodinka
Clinical and Vaccine Immunology.2015; 22(5): 484. CrossRef - NFAT1 and JunB Cooperatively Regulate IL-31 Gene Expression in CD4+ T Cells in Health and Disease
Ji Sun Hwang, Gi-Cheon Kim, EunBee Park, Jung-Eun Kim, Chang-Suk Chae, Won Hwang, Changhon Lee, Sung-Min Hwang, Hui Sun Wang, Chang-Duk Jun, Dipayan Rudra, Sin-Hyeog Im
The Journal of Immunology.2015; 194(4): 1963. CrossRef - Interleukin-31: A Novel Diagnostic Marker of Allergic Diseases
Anja Rabenhorst, Karin Hartmann
Current Allergy and Asthma Reports.2014;[Epub] CrossRef - Role of IL-31 in regulation of Th2 cytokine levels in patients with nasal polyps
Hong Ouyang, Jie Cheng, Yajun Zheng, Jingdong Du
European Archives of Oto-Rhino-Laryngology.2014; 271(10): 2703. CrossRef - Polymorphisms of interleukin-31 are associated with anti-CCP levels in females with rheumatoid arthritis
JI-IN YU, YOUNG-RAN PARK, SHIN-SEOK LEE, SOO-CHEON CHAE
Journal of Genetics.2014; 93(3): 813. CrossRef - IL-31 Associated with Coronary Artery Lesion Formation in Kawasaki Disease
Wan-Ning Tseng, Mao-Hung Lo, Mindy Ming-Huey Guo, Kai-Sheng Hsieh, Wei-Chiao Chang, Ho-Chang Kuo, Chien-Sheng Chen
PLoS ONE.2014; 9(8): e105195. CrossRef

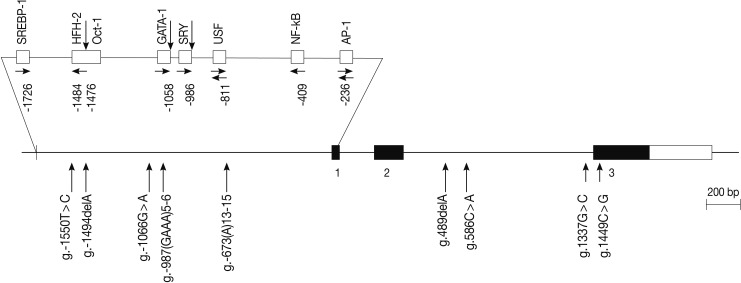
Fig. 1






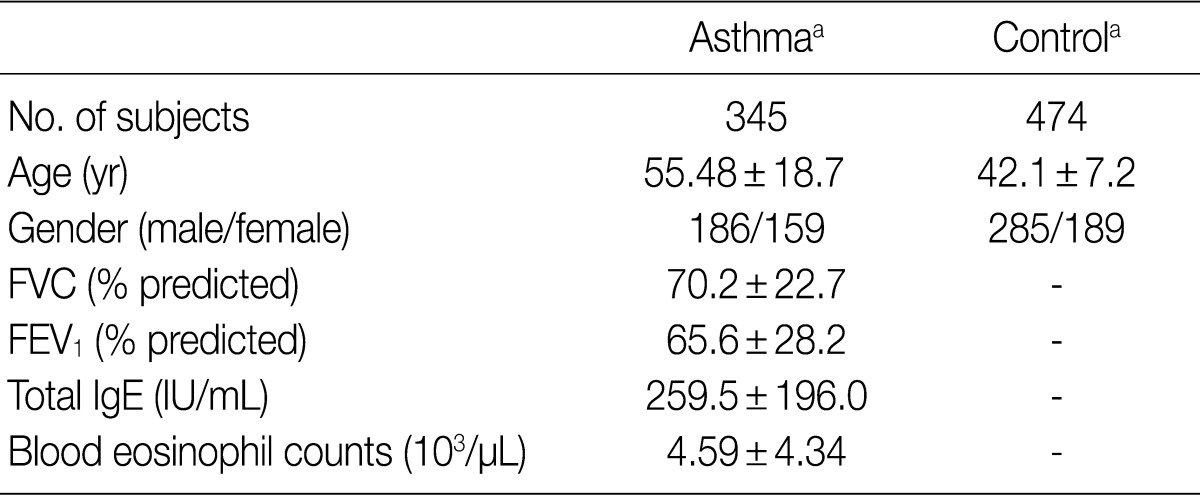
FVC, forced vital capacity; FEV1, forced expiratory volume in 1 sec. aValues are presented as means±standard deviation.
PCR, polymerase chain reaction;
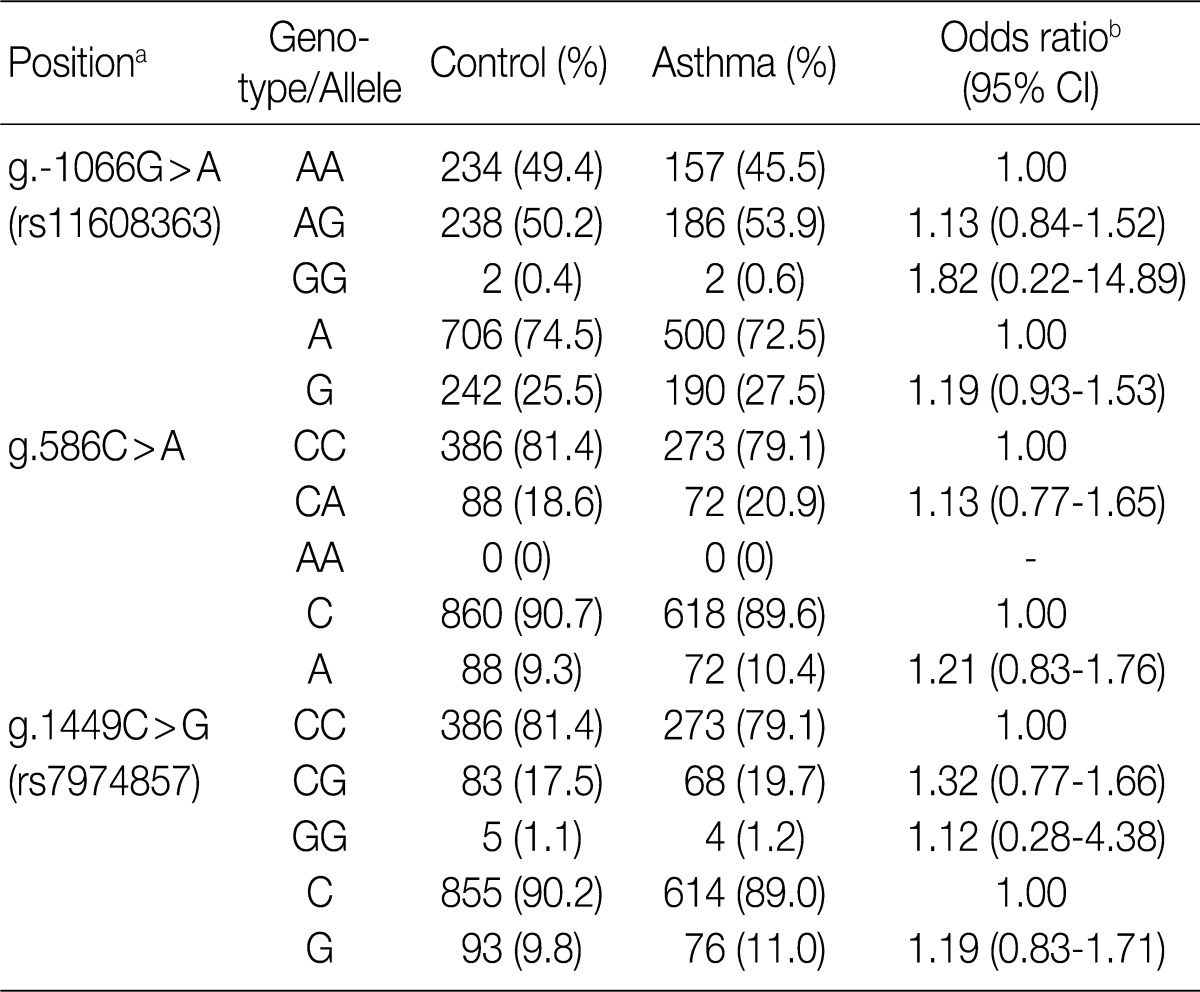
aCalculated from the translation start site; bLogistic regression analysis was used for calculating odds ratio (95% confidence interval [CI]), the results were adjusted for age.
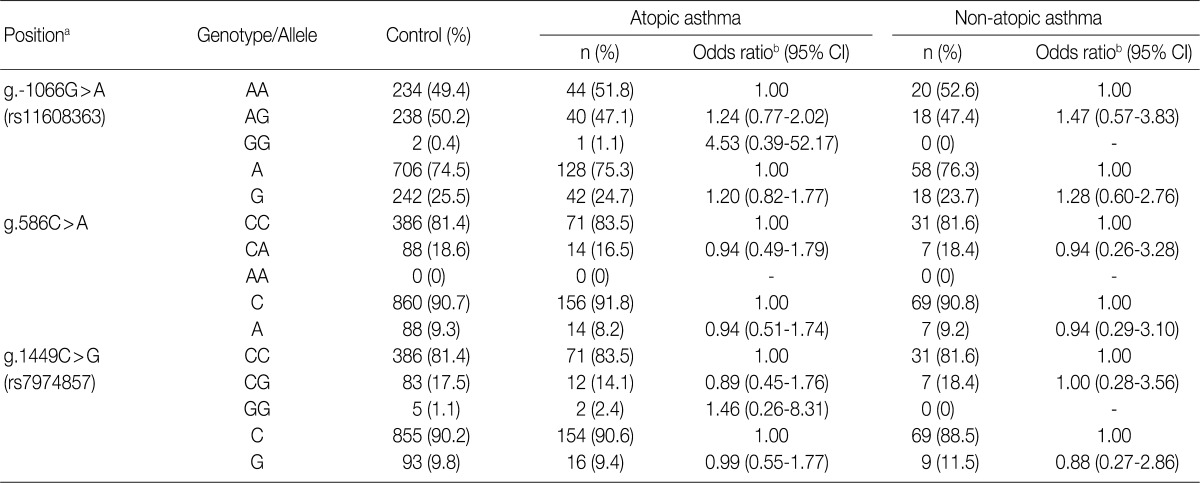
aCalculated from the translation start site; bLogistic regression analysis was used for calculating odds ratio (95% confidence interval [CI]), the results were adjusted for age.
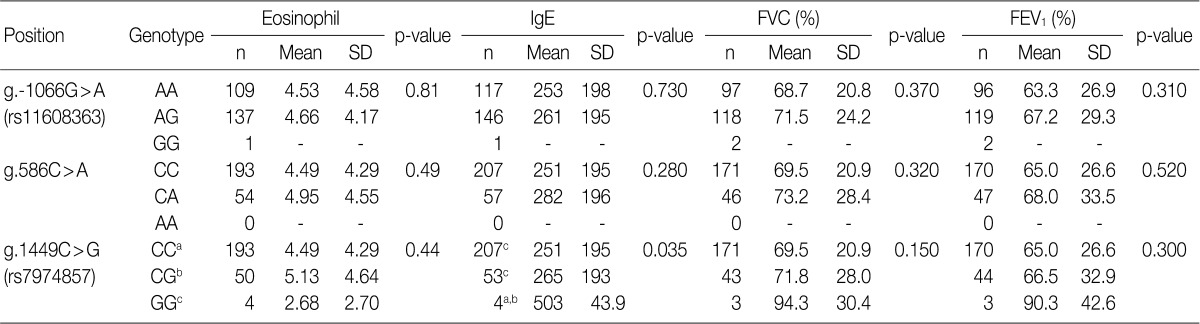
Data are analyzed by ANOVA. Significant differences (p<0.05) between two groups found by Bonferroni multiple comparisons are indicated by values which have the same letter (a-c). The position is calculated from the translation start site. FVC, forced vital capacity; FEV1, forced expiratory volume in 1 sec; SD, standard deviation.
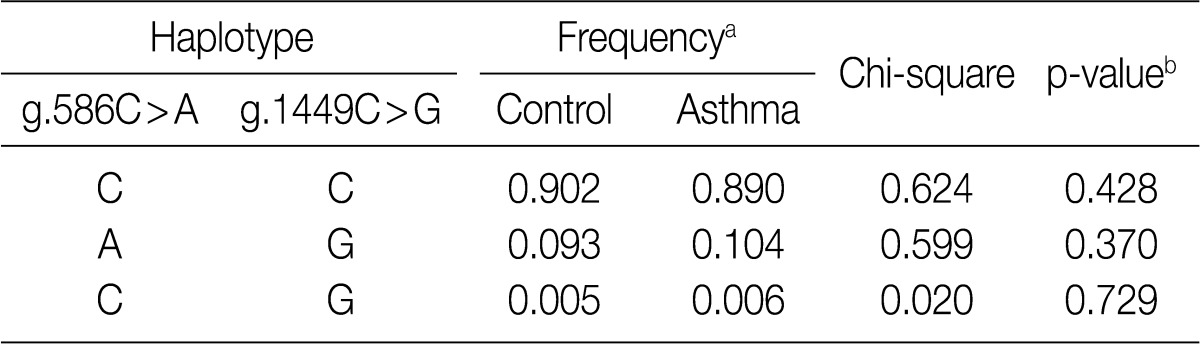
aValues are constructed by expectation-maximization algorithm with genotyped SNPs; bValues are analyzed by permutation test.

 E-submission
E-submission


 PubReader
PubReader Cite this Article
Cite this Article