Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 47(1); 2013 > Article
-
Original Article
Comparison of Direct Sequencing, PNA Clamping-Real Time Polymerase Chain Reaction, and Pyrosequencing Methods for the Detection ofEGFR Mutations in Non-small Cell Lung Carcinoma and the Correlation with Clinical Responses to EGFR Tyrosin - Hyun Ju Lee1,2, Xianhua Xu1, Hyojin Kim1, Yan Jin1, Pingli Sun1, Ji Eun Kim3, Jin-Haeng Chung1
-
Korean Journal of Pathology 2013;47(1):52-60.
DOI: https://doi.org/10.4132/KoreanJPathol.2013.47.1.52
Published online: February 25, 2013
1Department of Pathology, Seoul National University Bundang Hospital, Seoul National University College of Medicine, Seongnam, Korea.
2Department of Pathology, Soonchunhyang University Cheonan Hospital, Soonchunhyang University College of Medicine, Cheonan, Korea.
3Department of Pathology, Boramae Medical Center, Seoul National University College of Medicine, Seoul, Korea.
- Corresponding Author: Jin-Haeng Chung, M.D., Ph.D. Department of Pathology, Seoul National University Bundang Hospital, Seoul National University College of Medicine, 82 Gumi-ro 173beon-gil, Bundang-gu, Seongnam 463-707, Korea. Tel: +82-31-787-7713, Fax: +82-31-787-4012, chungjh@snu.ac.kr, Ji Eun Kim, M.D., Ph.D. Department of Pathology, Boramae Medical Center, Seoul National University College of Medicine, 20 Boramae-ro 5-gil, Dongjak-gu, Seoul 156-707, Korea. Tel: +82-2-870-2642, Fax: +82-2-831-0261, npol181@snu.ac.kr
© 2013 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- The aims of this study were to evaluate the abilities of direct sequencing (DS), peptide nucleic acid (PNA) clamping, and pyrosequencing methods to detect epidermal growth factor receptor (EGFR) mutations in formalin-fixed paraffin-embedded (FFPE) non-small cell lung carcinoma (NSCLC) samples and to correlate EGFR mutational status as determined by each method with the clinical response to EGFR tyrosine kinase inhibitors (TKIs).
-
Methods
- Sixty-one NSCLC patients treated with EGFR TKIs were identified to investigate somatic mutations in the EGFR gene (exons 18-21).
-
Results
- Mutations in the EGFR gene were detected in 38 of the 61 patients (62%) by DS, 35 (57%) by PNA clamping and 37 (61%) by pyrosequencing. A total of 44 mutations (72%) were found by at least one of the three methods, and the concordances among the results were relatively high (82-85%; kappa coefficient, 0.713 to 0.736). There were 15 discordant cases (25%) among the three different methods.
-
Conclusions
- All three EGFR mutation tests had good concordance rates (over 82%) for FFPE samples. These results suggest that if the DNA quality and enrichment of tumor cells are assured, then DS, PNA clamping, and pyrosequencing are appropriate methods for the detection of EGFR mutations.
- Patients and specimens
- Formalin-fixed paraffin-embedded (FFPE) tissues from 103 patients with NSCLC (26 biopsy and 77 resection samples) were obtained from the Seoul National University Bundang Hospital (SNUBH), Korea, between May 2003 and July 2010. All patients received EGFR TKIs gefitinib and erlotinib. We first analyzed 103 patients using DS and PNA clamping. However, 42 patients could not be tested by pyrosequencing either because the amount of available tissue was too small or because paraffin blocks were unavailable; consequently, 61 patients were included in this study. The patients included consisted of 26 men and 35 women. The mean age was 61.3 years (standard deviation, 10.6 years; range, 26 to 84 years), and the mean tumor size was 4.2 cm (standard deviation, 2.5 cm; range, 1.7 to 14.0 cm). All patients had undergone biopsy or surgical treatment (biopsy, n=1; wedge resection, n=1; lobectomy, n=56; and pneumonectomy, n=3).
- The hematoxylin and eosin-stained slides were independently reviewed by two pathologists (H.J.L. and J.H.C.) to confirm the original diagnosis of each patient based on the World Health Organization criteria.14
- The pathologic stage (p-stage) was determined at the time of the initial diagnosis using the 7th edition of the tumor-node-metastasis (TNM) classification.15 The stage Ia-IIIa patients had received EGFR TKIs when they relapsed. Patients were categorized as follows: never smokers (<100 lifetime cigarettes), former smokers (quit ≥1 year ago), or current smokers (quit <1 year ago). Additional data, including response, progression of the disease, survival status, and cause of death, were obtained from patients' medical records and/or through interviews with the families of the patients. The median follow-up period for all patients was 30.0 months, with a range of 2 to 111 months. All patient samples were tested with informed consent.
- DNA extraction
- Genomic DNA was extracted from FFPE tissue as described previously.16,17 A QIAamp DNA mini kit (Qiagen, Hilden, Germany) was used for genomic DNA isolation according to the manufacturer's instructions.
- Mutational analyses of EGFR genes: DS
- EGFR mutations in exons 18 to 21 were identified by nested polymerase chain reaction (PCR) and DS as described previously.16,17 All sequence variants were confirmed by sequencing the products of independent PCR amplifications in both directions. These sequences and chromatographs were manually compared with the EGFR reference sequence by two pathologists (H.J.L. and J.H.C.).
- Mutational analyses of EGFR genes: PNA clamping
- EGFR mutations were identified using the PNA Clamp EGFR mutation detection kit (Panagene, Daejeon, Korea) according to the manufacturer's instructions and as described previously.7-9 A CFX 384 real-time PCR instrument was used (Bio-Rad, Hercules, CA, USA). The threshold cycle (Ct) was automatically calculated from PCR amplification plots in which fluorescence was plotted against the number of cycles. Delta-Ct values were calculated as the Ct value from PCR with the PNA control minus the Ct value from PCR of the samples. A higher delta-Ct value indicates that the mutant was efficiently amplified. A cut-off value of 2 was used to differentiate the presence and absence of mutant DNA in the clinical samples.
- Mutational analyses of EGFR genes: pyrosequencing
- EGFR mutations in exons 18 to 21 were identified by pyrosequencing as described previously.12 An aliquot of 40 µL of PCR product was bound to streptavidin Sepharose HP (GE Healthcare, Uppsala, Sweden), purified, washed, denatured in 0.2 mol/L NaOH solution, and washed again. Then, 0.3 µmol/L of pyrosequencing primer was annealed to the purified single-stranded PCR product, and pyrosequencing was performed on a PyroMark ID system (Qiagen) following the manufacturer's instructions.
- Statistical analysis
- Statistical analyses were performed using SPSS ver. 17.0 (SPSS Inc., Chicago, IL, USA) Progression-free survival was assessed from the date of biopsy or surgical treatment to the earliest sign of disease progression, as determined using the Response Evaluation Criteria in Solid Tumors (RECIST), or to death from any cause.18 Overall survival was defined as the time from the date of biopsy or surgical treatment to the last follow-up visit or cancer-related death. Statistical significance was defined as p<0.05.
MATERIALS AND METHODS
- Comparison of the DS, PNA clamping, and pyrosequencing methods
- EGFR mutations were detected in 38 of the 61 patients (62%) by DS, in 35 (57%) using the PNA clamping method, and in 37 (61%) by pyrosequencing. There was good concordance (over 82%) in the assessment of EGFR mutations between DS and PNA clamping (concordant cases [n=50, 82%] and discordant cases [n=11, 18%]; kappa coefficient, 0.736), between DS and pyrosequencing (concordant cases [n=51, 84%] and discordant cases [n=10, 16%]; kappa coefficient, 0.716), and between PNA clamping and pyrosequencing (concordant cases [n=52, 85%] and discordant cases [n=9, 15%]; kappa coefficient, 0.713). Overall, there were concordant cases (n=46, 75%) and discordant cases (n=15, 25%) between the three different methods. In concordant cases, EGFR mutations were detected in 29 of the 61 patients (48%). A total of 44 mutations (72%) were found by at least one of the three methods.
- Among the discordant cases (Table 1), one patient (case no. 1) had at least one of the same mutations. In six patients (cases nos. 2-7), EGFR mutations were detected by PNA clamping or pyrosequencing but not by DS. For the four discordant cases (case no. 8-11) in which EGFR mutations were detected only by DS, the EGFR mutations were undesignated, and these patients had progressive disease (PD) when treated with TKIs. Table 2 presents detailed profiles of EGFR mutations identified by DS, not designed by PNA clamp EGFR mutation detection kit or pyrosequencing. In four cases (case nos. 12-15), an exon 21 mutation (L858R) was identified by DS, while the other two methods identified these as wild type in three cases (by PNA clamping) and in one case (by pyrosequencing).
- Among the concordant cases (Table 3), the TKI response (complete response [CR], partial response [PR], and stable disease [SD]) was significantly higher for the cases with exon 19 deletions or exon 21 point mutations than for those with wild-type EGFR (28% and 13% vs 4%). Of particular interest, an exon 19 deletion was identified in the one patient with squamous cell carcinoma (SCC) histology; this patient exhibited a PR. Seven patients with wild-type EGFR exhibited PD, four of whom had SCC and three of whom had other histological types.
- Patient characteristics and EGFR mutation status
- EGFR mutations detected by the three methods were found more frequently in female patients (71% by DS, 60% by PNA clamping, and 69% by pyrosequencing), never smokers (72% by DS, 61% by PNA clamping, and 67% by pyrosequencing), and adenocarcinoma (ADC) histology (70%, p=0.014, by DS; 68%, p<0.001, by PNA clamping; 70%, p=0.002, by pyrosequencing) (Table 4). Of particular interest, a significant number of male patients (50% by DS, 54% by PNA clamping, and 50% by pyrosequencing) and ever smokers (former and current smokers; 49% by DS, 46% by PNA clamping, and 46% by pyrosequencing) were found to have EGFR mutations by these three methods. Of the 61 patients, 1 had a CR, 13 had a PR, 14 had SD, and 33 had PD.
- Clinical outcomes among subgroups of patients treated with EGFR TKIs
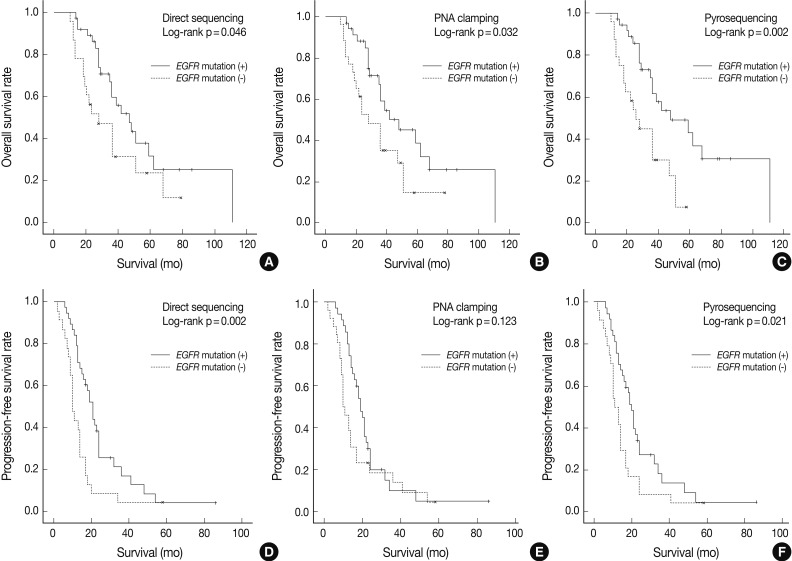
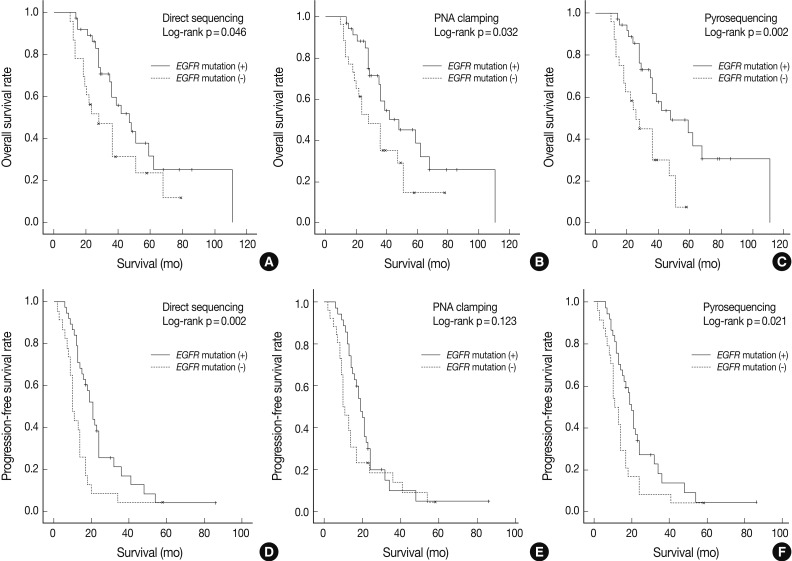
- As shown in Fig. 1, overall survival (p=0.046 for grouping by DS vs p=0.032 for PNA clamping vs p=0.002 for pyrosequencing) and progression-free survival (p=0.002 for grouping by DS vs p=0.021 for pyrosequencing) were significantly longer in patients with EGFR mutations detected by the three methods than in patients with wild-type EGFR.
- The objective response rate (OR) in the overall population was 23% (Table 5). The OR was significantly higher in the patients with EGFR mutations detected by pyrosequencing than in patients with wild-type EGFR (35% vs 4%, p=0.005). However, the number of samples was too low to make any definitive conclusions. The disease control rate (DCR) in the overall population was 46%. The DCR was higher in patients with ADC histology than in those with non-ADC histology (52% vs 19%, p=0.051), although this difference did not reach statistical significance. The DCR was significantly higher in the patients with EGFR mutations than in those with wild-type EGFR (p=0.019 for grouping by DS; p=0.001 for PNA clamping; p<0.001 for pyrosequencing).
- The median follow-up period for the analysis of progression-free survival was 16 months. The median progression-free survival was 19-20 months for the patients with EGFR mutations detected by the three methods and 10-12 months for those with wild-type EGFR (p=0.008 for grouping by DS; p=0.020 for PNA clamping; p=0.018 for pyrosequencing). The median follow-up period for the analysis of overall survival was 30 months. The median overall survival was 34-35 months for the patients with EGFR mutations detected by the three methods and 24-25 months for those with wild-type EGFR. These results suggest that EGFR mutations, not clinical predictors such as sex, smoking history, or histology, are associated with better outcomes with EGFR TKI treatment in terms of progression-free survival.
RESULTS
- In this study, three different methods for the detection of EGFR mutations-DS, PNA clamping, and pyrosequencing-were compared using samples from 61 NSCLC patients who were treated with EGFR TKIs. A total of 44 mutations (72%) were found by at least one of the three methods, and the concordances among the results were relatively high (82-85%; kappa coefficient, 0.713 to 0.736).
- Comparisons of DS with PNA clamping and pyrosequencing with respect to TKI responses have recently been published, and the results of those studies were quite different from our results.7,8,12,19 Kim et al.7 reported that EGFR mutations were detected in 63 of 240 NSCLC patients (26%) by DS, whereas PNA clamping detected EGFR mutations in 83 patients (35%). The patients in that study were from eight centers of the Korean Molecular Lung Cancer Group (KMLCG).7 The PNA clamping method was reported to detect 22 additional EGFR mutations-positive samples (10 in exon 19, 9 in exon 21, and 3 in both exons) and to identify more mutations than DS, although the clinical outcomes were not significantly different between the groups defined by each method. Dufort et al.12 reported that pyrosequencing is a highly accurate method for detecting EGFR mutations in patients with NSCLC. They found that three EGFR mutations-positive samples were detected only by pyrosequencing and not by DS, reflecting the lower sensitivity of the classical sequencing method.
- PNA is an artificially synthesized polymer that can bind to a complementary sequence in DNA; the binding capacity of PNA is stronger than that of DNA.7-9,20 The PNA clamping method is known to be more sensitive, rapid and simple to perform, and can detect mutant alleles even when present at levels 100-fold lower than wild-type alleles, whereas the minimum percentage of mutant DNA for analysis by DS is more than 25%. The minimum percentage of mutant DNA needed for analysis by pyrosequencing is at least 20%.12 However, the detection rates for EGFR mutations were not significantly different among the three methods used in this study. This lack of a significant difference might be due to 1) a higher proportion of tumor cells in the samples used in this study; 2) the macro- or microdissection of tumor cells prior to EGFR mutation tests by pathologists; and 3) the meticulous control of the turnaround time between the submission of the specimen to the pathology laboratory and formalin fixation in a single institution (SNUBH). During formalin fixation, the formaldehyde within tissues gradually changes to formic acid, which hydrolyzes DNA.6 DNA quality is affected by the fixation time and the type of fixative used.21 Greer et al.22 and Liu et al.23 suggested that tissues used for molecular tests should not be fixed for more than one day. To acquire a high proportion of tumor cells, a pathologist can, using a microscope, select an appropriate area from which DNA should be extracted. Thus, proper tissue handling (e.g., the timing of tissue sample acquisition, a shorter fixation time, and DNA quality control) by the pathologist is very important to improve the sensitivity of EGFR mutation tests.5 Goto et al.24 reported that it should be recognized that the detection rate of mutations by DS is largely influenced by the level of optimization in the processes implemented by the laboratory, and that the differences in reagents, DNA quality, software, and, crucially, primer design and amplicon size can affect the detection rate for DS. In particular, these researchers examined all the FFPE samples prepared by a single pathologist and generally found them to be of high quality and high tumor content.24
- In this study, 15 cases (25%) with discrepant results for the three methods were identified. EGFR mutations were detected in nine cases by DS, six cases by PNA clamping, and eight cases by pyrosequencing. Because PNA clamping can detect 29 designed target mutations of clinical significance among the approximately 250 EGFR mutations7 and because pyrosequencing has, in most cases, been used for the re-sequencing of a small number of selected hotspot codons,25 these two methods cannot detect undesignated EGFR mutation sites. Four out of the 15 discrepant cases (cases nos. 8-11) (Table 2) were identified only by DS. The mutations in these cases were undesignated EGFR mutations and were thus not detectable by PNA clamping or pyrosequencing. The clinical significance of these rare mutations is still uncertain, and further analyses are needed. The weakness of PNA clamping and pyrosequencing is that these methods can only be used to detect mutations for which primers have been individually designed; in contrast, DS is able to uncover novel mutations.
- In six patients (cases nos. 2-7) (Table 1), EGFR mutations were detected only by PNA clamping or pyrosequencing, but not by DS. These cases might have had less than 25% mutant DNA in the tested sample, therefore making these EGFR mutations undetectable by DS.
- The types of EGFR mutations identified in this study were in accordance with those found in previous studies.2,26 The most frequent mutations were an in-frame deletion in exon 19 (53-54%) and the L858R point mutation in exon 21 (35-39%); these mutations accounted for over 90% of the detected mutations. Patients who harbored activating EGFR mutations showed a positive clinical response to EGFR TKIs; the OR for these patients was 23% and the DCR was 46%. The OR and DCR were higher in EGFR-mutation-positive patients than in patients with wild-type EGFR, as reported in previous retrospective and prospective studies.27 Only mutational analysis by pyrosequencing was statistically significantly correlated with OR. However, the number of samples was too low to make any definitive conclusions.
- Mutational analysis by DS, PNA clamping, and pyrosequencing was successful and confirmed a strong and independent association between EGFR mutations and clinical outcome. As expected, EGFR mutations were more frequent in women who had never smoked and in those with ADC histology. In addition, a significant number of males (51%) and ever smokers (47%, current and former smokers) were found to have EGFR mutations by the three methods. Thus, it is necessary to test for EGFR mutations not only in female never smokers but also in males and ever smokers. Clinical predictors, such as sex, smoking history, and histology, added little predictive information to that provided by the mutational analysis. These data indicate that the mutational status of EGFR is the most important predictor of clinical outcome in EGFR TKIs-treated patients.17
- In conclusion, all three EGFR mutation tests had good concordance rates (over 82%) for FFPE samples. These results suggest that if the DNA quality and enrichment of tumor cells are assured, then DS, PNA clamping, and pyrosequencing are appropriate methods for the detection of EGFR mutations. The presence of some cases with discordant results for the three different methods indicates that these methods must be further standardized and validated.
DISCUSSION
Acknowledgments
Acknowledgments
- 1. Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304: 1497-1500. ArticlePubMed
- 2. Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129-2139. ArticlePubMed
- 3. Penzel R, Sers C, Chen Y, et al. EGFR mutation detection in NSCLC: assessment of diagnostic application and recommendations of the German Panel for Mutation Testing in NSCLC. Virchows Arch 2011; 458: 95-98. ArticlePubMedPDF
- 4. Helland Å, Skaug HM, Kleinberg L, et al. EGFR gene alterations in a Norwegian cohort of lung cancer patients selected for surgery. J Thorac Oncol 2011; 6: 947-950. ArticlePubMed
- 5. Pirker R, Herth FJ, Kerr KM, et al. Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol 2010; 5: 1706-1713. ArticlePubMed
- 6. Eberhard DA, Giaccone G, Johnson BE. Non-Small-Cell Lung Cancer Working Group. Biomarkers of response to epidermal growth factor receptor inhibitors in Non-Small-Cell Lung Cancer Working Group: standardization for use in the clinical trial setting. J Clin Oncol 2008; 26: 983-994. ArticlePubMed
- 7. Kim HJ, Lee KY, Kim YC, et al. Detection and comparison of peptide nucleic acid-mediated real-time polymerase chain reaction clamping and direct gene sequencing for epidermal growth factor receptor mutations in patients with non-small cell lung cancer. Lung Cancer 2012; 75: 321-325. ArticlePubMed
- 8. Kim HJ, Kim WS, Shin KC, et al. Comparative analysis of peptide nucleic acid (PNA)-mediated real-time PCR clamping and DNA direct sequencing for EGFR mutation detection. Tuberc Respir Dis 2011; 70: 21-27. Article
- 9. Lee KY, Kim HJ, Kim SJ, et al. PNA-mediated PCR clamping for the detection of EGFR mutations in non-small cell lung cancer. Tuberc Respir Dis 2010; 69: 271-278. Article
- 10. Ronaghi M, Uhlén M, Nyrén P. A sequencing method based on real-time pyrophosphate. Science 1998; 281: 363365.ArticlePubMed
- 11. Takano T, Ohe Y, Sakamoto H, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol 2005; 23: 6829-6837. ArticlePubMed
- 12. Dufort S, Richard MJ, Lantuejoul S, de Fraipont F. Pyrosequencing, a method approved to detect the two major EGFR mutations for anti EGFR therapy in NSCLC. J Exp Clin Cancer Res 2011; 30: 57.ArticlePubMedPMCPDF
- 13. Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res 2007; 13: 4954-4955. ArticlePubMedPDF
- 14. Travis WD, Brambilla E, Müller-Hermelink HK, Harris CC. World Health Organization classification of tumours: pathology and genetics of tumours of the lung, pleura, thymus and heart. 2004; Lyon: IARC Press.
- 15. Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol 2007; 2: 706-714. ArticlePubMed
- 16. Yoo SB, Chung JH, Lee HJ, Lee CT, Jheon S, Sung SW. Epidermal growth factor receptor mutation and p53 overexpression during the multistage progression of small adenocarcinoma of the lung. J Thorac Oncol 2010; 5: 964-969. ArticlePubMed
- 17. Sun PL, Seol H, Lee HJ, et al. High incidence of EGFR mutations in Korean men smokers with no intratumoral heterogeneity of lung adenocarcinomas: correlation with histologic subtypes, EGFR/TTF-1 expressions, and clinical features. J Thorac Oncol 2012; 7: 323-330. ArticlePubMed
- 18. Therasse P, Arbuck SG, Eisenhauer EA, et al. European Organization for Research and Treatment of Cancer. National Cancer Institute of the United States. National Cancer Institute of Canada. New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst 2000; 92: 205-216. ArticlePubMed
- 19. Han HS, Lim SN, An JY, et al. Detection of EGFR mutation status in lung adenocarcinoma specimens with different proportions of tumor cells using two methods of differential sensitivity. J Thorac Oncol 2012; 7: 355-364. ArticlePubMed
- 20. Choi JJ, Kim C, Park H. Peptide nucleic acid-based array for detecting and genotyping human papillomaviruses. J Clin Microbiol 2009; 47: 1785-1790. ArticlePubMedPMCPDF
- 21. Seo AN, Kim JH, Lee D, Jeong JY, Park JY. Comparison of the DNA preservation in neutral-buffered formalin fixed paraffin-embedded tissue and in non-buffered formalin fixed paraffin-embedded tissue. Korean J Pathol 2011; 45: 549-556. Article
- 22. Greer CE, Wheeler CM, Manos MM. Sample preparation and PCR amplification from paraffin-embedded tissues. PCR Methods Appl 1994; 3: S113-S122. ArticlePubMed
- 23. Liu D, Nakano J, Ueno M, et al. A useful protocol for analyses of mutations of the epidermal growth factor receptor gene. Oncol Rep 2006; 15: 1503-1505. ArticlePubMed
- 24. Goto K, Satouchi M, Ishii G, et al. An evaluation study of EGFR mutation tests utilized for non-small-cell lung cancer in the diagnostic setting. Ann Oncol 2012; 23: 2914-2919. ArticlePubMed
- 25. Ahmadian A, Ehn M, Hober S. Pyrosequencing: history, biochemistry and future. Clin Chim Acta 2006; 363: 83-94. ArticlePubMed
- 26. Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009; 361: 947-957. ArticlePubMed
- 27. Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol 2005; 23: 2513-2520. ArticlePubMed
REFERENCES
EGFR, epidermal growth factor receptor; PNA, peptide nucleic acid; TKI, tyrosine kianse inhibitor; SD, stable disease; F, female; N, never smoker; ADC, adenocarcinoma; PR, partial response; M, male; C, current smoker; ASC, adenosquamous cell carcinoma; PD, progressive disease; FS, former smoker; LCC, large cell carcinoma.
EGFR, epidermal growth factor receptor; PNA, peptide nucleic acid; TKI, tyrosine kianse inhibitor; CR, complete response; F, female; N, never smoker; ADC, adenocarcinoma; PR, partial response; SCC, squamous cell carcinoma; M, male; C, current smoker; SD, stable disease; FS, former smoker; PD, progressive disease; LCNEC, large cell neuroendocrine carcinoma; SarCa, sarcomatoid carcinoma.
| n (%) |
Direct sequencing EGFR mutation |
PNA clamping EGFR mutation |
Pyrosequencing EGFR mutation |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (+) | (-) | p-value | (+) | (-) | p-value | (+) | (-) | p-value | ||
| Total | 61 (100) | 38 (62) | 23 (38) | 35 (57) | 26 (43) | 37 (61) | 24 (39) | |||
| Sex | ||||||||||
| Female | 35 (57) | 25 (71) | 10 (29) | 0.113 | 21 (60) | 14 (40) | 0.794 | 24 (69) | 11 (31) | 0.188 |
| Male | 26 (43) | 13 (50) | 13 (50) | 14 (54) | 12 (46) | 13 (50) | 13 (50) | |||
| Smoking historya | ||||||||||
| Never | 36 (59) | 26 (72) | 10 (28) | 0.066b | 22 (61) | 14 (39) | 0.600b | 24 (67) | 12 (33) | 0.294b |
| Former | 6 (10) | 3 (50) | 3 (50) | 2 (33) | 4 (67) | 2 (33) | 4 (67) | |||
| Current | 19 (31) | 9 (47) | 10 (53) | 11 (58) | 8 (42) | 11 (58) | 8 (42) | |||
| Histology | ||||||||||
| ADC | 50 (82) | 35 (70) | 15 (30) | 0.014c | 34 (68) | 16 (32) | <0.001c | 35 (70) | 15 (30) | 0.002c |
| SCC | 5 (8) | 1 (20) | 4 (80) | 1 (20) | 4 (80) | 1 (20) | 4 (80) | |||
| Others | 6 (10) | 2 (33) | 4 (67) | 0 | 6 (100) | 1 (17) | 5 (83) | |||
| Tumor size (cm) | ||||||||||
| ≤ 3 | 26 (43) | 21 (81) | 5 (19) | 0.016 | 19 (73) | 7 (27) | 0.040 | 21 (81) | 5 (19) | 0.008 |
| > 3 | 35 (57) | 17 (49) | 18 (51) | 16 (46) | 19 (54) | 16 (46) | 19 (54) | |||
| Operation method | ||||||||||
| Biopsy | 1 (2) | 1 (100) | 0 | 1.000 | 1 (100) | 0 | 1.000 | 1 (100) | 0 | 1.000 |
| Resection | 60 (98) | 37 (62) | 23 (38) | 34 (57) | 26 (43) | 36 (60) | 24 (40) | |||
| EGFR TKI response | ||||||||||
| CR | 1 (2) | 1 (100) | 0 | 0.537d | 1 (100) | 0 | 0.122d | 1 (100) | 0 | 0.005d |
| PR | 13 (21) | 9 (69) | 4 (31) | 10 (77) | 3 (23) | 12 (92) | 1 (8) | |||
| SD | 14 (23) | 12 (86) | 2 (14) | 12 (86) | 2 (14) | 12 (86) | 2 (14) | |||
| PD | 33 (54) | 16 (48) | 17 (52) | 12 (36) | 21 (64) | 12 (36) | 21 (64) | |||
EGFR, epidermal growth factor receptor; PNA, peptide nucleic acid; ADC, adenocarcinoma; SCC, squamous cell carcinoma; TKI, tyrosine kianse inhibitor; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease.
aNever smokers were defined as patients who had a lifetime smoking exposure of <100 cigarettes and former smokers were defined as patients who had stopped smoking at least 1 yr before diagnosis;
bComparison between never smokers and others;
cComparison between adenocarcinoma and nonadenocarcinoma;
dComparison between CR, PR and SD, PD.
| n (%) | Objective responsea | p-value | Disease control rateb | p-value | Median TTP (mo) | p-value | MST (mo) | p-value | |
|---|---|---|---|---|---|---|---|---|---|
| Total | 61 (100) | 14 (23) | 28 (46) | 16.0 | 30.0 | ||||
| Sex | |||||||||
| Female | 35 (57) | 7 (20) | 0.553 | 15 (43) | 0.613 | 18.0 | 0.440 | 29.0 | 0.445 |
| Male | 26 (43) | 7 (27) | 13 (50) | 13.5 | 35.0 | ||||
| Smoking historyc | |||||||||
| Never | 36 (59) | 9 (25) | 0.762d | 17 (47) | 1.000d | 17.0 | 0.300d | 32.0 | 0.795d |
| Former | 6 (10) | 1 (17) | 3 (50) | 19.5 | 44.5 | ||||
| Current | 19 (31) | 4 (21) | 8 (42) | 11.0 | 28.0 | ||||
| Histology | |||||||||
| ADC | 50 (82) | 12 (24) | 0.726e | 26 (52) | 0.051e | 17.5 | 0.043e | 30.0 | 0.100e |
| SCC | 5 (8) | 1 (20) | 1 (20) | 11.0 | 28.0 | ||||
| Others | 6 (10) | 1 (17) | 1 (17) | 10.0 | 28.0 | ||||
| Direct sequencing | |||||||||
| EGFR mutation (+) | 38 (62) | 10 (26) | 0.537 | 22 (58) | 0.019 | 20.0 | 0.008 | 34.5 | 0.428 |
| EGFR mutation (-) | 23 (38) | 4 (17) | 6 (26) | 10.0 | 24.0 | ||||
| PNA clamping | |||||||||
| EGFR mutation (+) | 35 (57) | 11 (31) | 0.122 | 23 (66) | 0.001 | 19.0 | 0.020 | 34.0 | 0.606 |
| EGFR mutation (-) | 26 (43) | 3 (12) | 5 (23) | 10.5 | 24.0 | ||||
| Pyrosequencing | |||||||||
| EGFR mutation (+) | 37 (61) | 13 (35) | 0.005 | 25 (68) | <0.001 | 19.0 | 0.018 | 35.0 | 0.294 |
| EGFR mutation (-) | 24 (39) | 1 (4) | 3 (13) | 12.0 | 25.0 |
EGFR, epidermal growth factor receptor; TKI, tyrosine kianse inhibitor; TTP, time-to-progression; MST, median survival time; ADC, adenocarcinoma; SCC, squamous cell carcinoma.
aObjective response: complete response or partial response;
bDisease control rate: complete response or partial response or stable disease;
cNever smokers were defined as patients who had a lifetime smoking exposure of <100 cigarettes and former smokers were defined as patients who had stopped smoking at least 1 yr before diagnosis;
dComparison between never smokers and others;
eComparison between adenocarcinoma and nonadenocarcinoma.
Figure & Data
References
Citations

- Recent Trends of Lung Cancer in Korea
Jae Guk Lee, Ho Cheol Kim, Chang-Min Choi
Tuberculosis and Respiratory Diseases.2021; 84(2): 89. CrossRef - Predictive value of KRAS mutation and excision repair cross-complementing 1 (ERCC1) protein overexpression in patients with colorectal cancer administered FOLFOX regimen
Sun Min Park, Sung Bong Choi, Yoon Suk Lee, In Kyu Lee
Asian Journal of Surgery.2021; 44(5): 715. CrossRef - Recent advances in diagnostic technologies in lung cancer
Hye Jung Park, Sang Hoon Lee, Yoon Soo Chang
The Korean Journal of Internal Medicine.2020; 35(2): 257. CrossRef - Afatinib is effective in the treatment of lung adenocarcinoma with uncommon EGFR p.L747P and p.L747S mutations
Sheng-Kai Liang, Jen-Chung Ko, James Chih-Hsin Yang, Jin-Yuan Shih
Lung Cancer.2019; 133: 103. CrossRef - Biomarker Testing for Patients With Advanced Non–Small Cell Lung Cancer: Real-World Issues and Tough Choices
Nathan A. Pennell, Maria E. Arcila, David R. Gandara, Howard West
American Society of Clinical Oncology Educational Book.2019; (39): 531. CrossRef - Evaluation of EGFR mutations in NSCLC with highly sensitive droplet digital PCR assays
Xi‑Wen Jiang, Wei Liu, Xiao‑Ya Zhu, Xiao‑Xie Xu
Molecular Medicine Reports.2019;[Epub] CrossRef - Peptide Nucleic Acid Clamping and Direct Sequencing in the Detection of Oncogenic Alterations in Lung Cancer: Systematic Review and Meta-Analysis
Jae-Uk Song, Jonghoo Lee
Yonsei Medical Journal.2018; 59(2): 211. CrossRef - Distribution of KRAS, DDR2, and TP53 gene mutations in lung cancer: An analysis of Iranian patients
Zahra Fathi, Seyed Ali Javad Mousavi, Raheleh Roudi, Farideh Ghazi, Sumitra Deb
PLOS ONE.2018; 13(7): e0200633. CrossRef - EGFR T790M mutation testing within the osimertinib AURA Phase I study
Simon Dearden, Helen Brown, Suzanne Jenkins, Kenneth S. Thress, Mireille Cantarini, Rebecca Cole, Malcolm Ranson, Pasi A. Jänne
Lung Cancer.2017; 109: 9. CrossRef - Molecular Testing of Lung Cancers
Hyo Sup Shim, Yoon-La Choi, Lucia Kim, Sunhee Chang, Wan-Seop Kim, Mee Sook Roh, Tae-Jung Kim, Seung Yeon Ha, Jin-Haeng Chung, Se Jin Jang, Geon Kook Lee
Journal of Pathology and Translational Medicine.2017; 51(3): 242. CrossRef - Mutations of the Epidermal Growth Factor Receptor Gene in Triple-Negative Breast Cancer
Aeri Kim, Min Hye Jang, Soo Jung Lee, Young Kyung Bae
Journal of Breast Cancer.2017; 20(2): 150. CrossRef - Double primary lung adenocarcinoma diagnosed by epidermal growth factor receptor mutation status
Oh Jung Kwon, Min Hyeok Lee, Sung Ju Kang, Seul Gi Kim, In Beom Jeong, Ji Yun Jeong, Eun Jung Cha, Do Yeun Cho, Young Jin Kim, Ji Woong Son
Yeungnam University Journal of Medicine.2017; 34(2): 270. CrossRef - Generation of lung cancer cell lines harboring EGFR T790M mutation by CRISPR/Cas9-mediated genome editing
Mi-Young Park, Min Hee Jung, Eun Young Eo, Seokjoong Kim, Sang Hoon Lee, Yeon Joo Lee, Jong Sun Park, Young Jae Cho, Jin Haeng Chung, Cheol Hyeon Kim, Ho Il Yoon, Jae Ho Lee, Choon-Taek Lee
Oncotarget.2017; 8(22): 36331. CrossRef - Comparison of EGFR mutation detection between the tissue and cytology using direct sequencing, pyrosequencing and peptide nucleic acid clamping in lung adenocarcinoma: Korean multicentre study
Kyueng-Whan Min, Wan-Seop Kim, Se Jin Jang, Yoo Duk Choi, Sunhee Chang, Soon Hee Jung, Lucia Kim, Mee Sook Roh, Choong Sik Lee, Jung Weon Shim, Mi Jin Kim, Geon Kook Lee
QJM.2016; 109(3): 167. CrossRef - Epidermal Growth Factor Receptor Mutation and Anaplastic Lymphoma Kinase Gene Fusion: Detection in Malignant Pleural Effusion by RNA or PNA Analysis
Yi-Lin Chen, Chung-Ta Lee, Cheng-Chan Lu, Shu-Ching Yang, Wan-Li Chen, Yang-Cheng Lee, Chung-Hsien Yang, Shu-Ling Peng, Wu-Chou Su, Nan-Haw Chow, Chung-Liang Ho, Javier S Castresana
PLOS ONE.2016; 11(6): e0158125. CrossRef - IDH Mutation Analysis in Ewing Sarcoma Family Tumors
Ki Yong Na, Byeong-Joo Noh, Ji-Youn Sung, Youn Wha Kim, Eduardo Santini Araujo, Yong-Koo Park
Journal of Pathology and Translational Medicine.2015; 49(3): 257. CrossRef - Immunohistochemical demonstration of alteration of β-catenin during tumor metastasis by different mechanisms according to histology in lung cancer
XIANHUA XU, JI EUN KIM, PING-LI SUN, SEOL BONG YOO, HYOJIN KIM, YAN JIN, JIN-HAENG CHUNG
Experimental and Therapeutic Medicine.2015; 9(2): 311. CrossRef - Detection of EGFR-TK Domain–activating Mutations in NSCLC With Generic PCR-based Methods
Rajendra B. Shahi, Sylvia De Brakeleer, Jacques De Grève, Caroline Geers, Peter In’t Veld, Erik Teugels
Applied Immunohistochemistry & Molecular Morphology.2015; 23(3): 163. CrossRef - Frequent aerogenous spread with decreased E-cadherin expression of ROS1- rearranged lung cancer predicts poor disease-free survival
Yan Jin, Ping-Li Sun, Soo Young Park, Hyojin Kim, Eunhyang Park, Gilhyang Kim, Sukki Cho, Kwhanmien Kim, Choon-Taek Lee, Jin-Haeng Chung
Lung Cancer.2015; 89(3): 343. CrossRef - Membranous Insulin-like Growth Factor-1 Receptor (IGF1R) Expression Is Predictive of Poor Prognosis in Patients with Epidermal Growth Factor Receptor (EGFR)-Mutant Lung Adenocarcinoma
Eunhyang Park, Soo Young Park, Hyojin Kim, Ping-Li Sun, Yan Jin, Suk Ki Cho, Kwhanmien Kim, Choon-Taek Lee, Jin-Haeng Chung
Journal of Pathology and Translational Medicine.2015; 49(5): 382. CrossRef - Peptide Nucleic Acid Clamping Versus Direct Sequencing for the Detection of EGFR Gene Mutation in Patients with Non-small Cell Lung Cancer
Seong-Hoon Yoon, Yoo-Duk Choi, In-Jae Oh, Kyu-Sik Kim, Hayoung Choi, Jinsun Chang, Hong-Joon Shin, Cheol-Kyu Park, Young-Chul Kim
Cancer Research and Treatment.2015; 47(4): 661. CrossRef - Analysis of Mutations in Epidermal Growth Factor Receptor Gene in Korean Patients with Non-small Cell Lung Cancer: Summary of a Nationwide Survey
Sang Hwa Lee, Wan Seop Kim, Yoo Duk Choi, Jeong Wook Seo, Joung Ho Han, Mi Jin Kim, Lucia Kim, Geon Kook Lee, Chang Hun Lee, Mee Hye Oh, Gou Young Kim, Sun Hee Sung, Kyo Young Lee, Sun Hee Chang, Mee Sook Rho, Han Kyeom Kim, Soon Hee Jung, Se Jin Jang
Journal of Pathology and Translational Medicine.2015; 49(6): 481. CrossRef - Novel EGFR mutation-specific antibodies for lung adenocarcinoma: Highly specific but not sensitive detection of an E746_A750 deletion in exon 19 and an L858R mutation in exon 21 by immunohistochemistry
An Na Seo, Tae-In Park, Yan Jin, Ping-Li Sun, Hyojin Kim, Hyun Chang, Jin-Haeng Chung
Lung Cancer.2014; 83(3): 316. CrossRef - Simultaneous diagnostic platform of genotyping EGFR, KRAS, and ALK in 510 Korean patients with non‐small‐cell lung cancer highlights significantly higher ALK rearrangement rate in advanced stage
Tae‐Jung Kim, Chan Kwon Park, Chang Dong Yeo, Kihoon Park, Chin Kook Rhee, Jusang Kim, Seung Joon Kim, Sang Haak Lee, Kyo‐Young Lee, Hyoung‐Kyu Yoon
Journal of Surgical Oncology.2014; 110(3): 245. CrossRef - Epidermal growth factor receptor mutations and anaplastic lymphoma kinase rearrangements in lung cancer with nodular ground-glass opacity
Sung-Jun Ko, Yeon Joo Lee, Jong Sun Park, Young-Jae Cho, Ho Il Yoon, Jin-Haeng Chung, Tae Jung Kim, Kyung Won Lee, Kwhanmien Kim, Sanghoon Jheon, Hyojin Kim, Jae Ho Lee, Choon-Taek Lee
BMC Cancer.2014;[Epub] CrossRef - Cytoplasmic YAP Expression is Associated with Prolonged Survival in Patients with Lung Adenocarcinomas and Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Treatment
Ping-Li Sun, Ji Eun Kim, Seol Bong Yoo, Hyojin Kim, Yan Jin, Sanghoon Jheon, Kwhanmien Kim, Choon Taek Lee, Jin-Haeng Chung
Annals of Surgical Oncology.2014; 21(S4): 610. CrossRef - Sensitive methods for detection of the S768R substitution in exon 18 of the DDR2 gene in patients with central nervous system metastases of non-small cell lung cancer
Marcin Nicoś, Tomasz Powrózek, Paweł Krawczyk, Bożena Jarosz, Beata Pająk, Marek Sawicki, Krzysztof Kucharczyk, Tomasz Trojanowski, Janusz Milanowski
Medical Oncology.2014;[Epub] CrossRef - Clinicopathologic and prognostic significance of c-MYC copy number gain in lung adenocarcinomas
A N Seo, J M Yang, H Kim, S Jheon, K Kim, C T Lee, Y Jin, S Yun, J-H Chung, J H Paik
British Journal of Cancer.2014; 110(11): 2688. CrossRef - KRASMutation Detection in Non-small Cell Lung Cancer Using a Peptide Nucleic Acid-Mediated Polymerase Chain Reaction Clamping Method and Comparative Validation with Next-Generation Sequencing
Boram Lee, Boin Lee, Gangmin Han, Mi Jung Kwon, Joungho Han, Yoon-La Choi
Korean Journal of Pathology.2014; 48(2): 100. CrossRef - Guideline Recommendations forEGFRMutation Testing in Lung Cancer: Proposal of the Korean Cardiopulmonary Pathology Study Group
Hyo Sup Shim, Jin-Haeng Chung, Lucia Kim, Sunhee Chang, Wan-Seop Kim, Geon Kook Lee, Soon-Hee Jung, Se Jin Jang
Korean Journal of Pathology.2013; 47(2): 100. CrossRef - Immunohistochemical Classification of Primary and Secondary Glioblastomas
Kyu Sang Lee, Gheeyoung Choe, Kyung Han Nam, An Na Seo, Sumi Yun, Kyung Ju Kim, Hwa Jin Cho, Sung Hye Park
Korean Journal of Pathology.2013; 47(6): 541. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure

Fig. 1
| Case No. | Direct sequencing | PNA clamping | Pyrosequencing | TKI Response | Sex | Smoking | Histology |
|---|---|---|---|---|---|---|---|
| 1 | G719X, L861Q | G719X, L861Q | G719X | SD | F | N | ADC |
| 2 | Wild | Wild | Exon 19 del | PR | M | C | ASC |
| 3 | Wild | Wild | Exon 19 del | PD | F | N | ADC |
| 4 | Wild | G719X | Wild | SD | M | C | ADC |
| 5 | Wild | Exon 19 del | Exon 19 del | PR | M | C | ADC |
| 6 | Wild | S768I | Wild | PD | F | N | ADC |
| 7 | Wild | L858R | L858R | PR | M | N | ADC |
| 8 | Exon 19 del | Wild | Wild | PD | F | N | ADC |
| 9 | Exon 19 del | Wild | Wild | PD | M | FS | ASC |
| 10 | Exon 20 duplication | Wild | Wild | PD | F | N | ADC |
| 11 | R776H | Wild | Wild | PD | M | N | LCC |
| 12 | L858R | L858R | Wild | PD | M | N | ADC |
| 13 | L858R | Wild | L858R | PR | F | N | ADC |
| 14 | L858R | Wild | L858R | SD | F | N | ADC |
| 15 | L858R | Wild | L858R | PD | F | N | ADC |
| Case No. | Exon | Alteration | Direct sequencing |
TKI response | |
|---|---|---|---|---|---|
| Nucleotide alteration | Amino acid alteration | ||||
| 8 | 19 | Deletion | 2239-2263del | L747-755Adel | PD |
| 9 | 19 | Deletion | 2253-2276del | S752_I759del | PD |
| 10 | 20 | Duplication | dup 2311-2319 AACCCCCAC | D770_N771insNPH | PD |
| 11 | 20 | Point mutation | 2327G>A | R776H | PD |
| EGFR mutation | TKI response | Sex | Smoking | Histology |
|---|---|---|---|---|
| Exon19 (n = 18, 29%) | CR (n = 2,3%) | F | N | ADC |
| PR (n = 3) | F | N | ADC | |
| F | N | SCC | ||
| M | C | ADC | ||
| SD (n = 9) | F (n = 5) | N | ADC | |
| M (n = 2) | FS | ADC | ||
| M (n = 2) | C | ADC | ||
| PD (n = 5) | F (n = 3) | N | ADC | |
| M (n = 2) | C | ADC | ||
| Exon21 (n = 11, 18%) | PR (n = 5) | F (n = 3) | N | ADC |
| M | N | ADC | ||
| M | C | ADC | ||
| SD | M | C | ADC | |
| PD (n = 5) | F (n = 3) | N | ADC | |
| F (n = 2) | C | ADC | ||
| Wild (n = 17, 28%) | PR | M | FS | ADC |
| SD (n = 1,3%) | F | N | ADC | |
| PD (n = 15) | F (n = 4) | N | ADC | |
| F | N | SCC | ||
| F | C | ADC | ||
| F | C | LCNEC | ||
| M | N | SCC | ||
| M (n = 2) | FS | ADC | ||
| M | C | ADC | ||
| M (n = 2) | C | SCC | ||
| M (n = 2) | C | SarCa |
| n (%) | Direct sequencing EGFR mutation |
PNA clamping EGFR mutation |
Pyrosequencing EGFR mutation |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (+) | (-) | p-value | (+) | (-) | p-value | (+) | (-) | p-value | ||
| Total | 61 (100) | 38 (62) | 23 (38) | 35 (57) | 26 (43) | 37 (61) | 24 (39) | |||
| Sex | ||||||||||
| Female | 35 (57) | 25 (71) | 10 (29) | 0.113 | 21 (60) | 14 (40) | 0.794 | 24 (69) | 11 (31) | 0.188 |
| Male | 26 (43) | 13 (50) | 13 (50) | 14 (54) | 12 (46) | 13 (50) | 13 (50) | |||
| Smoking history |
||||||||||
| Never | 36 (59) | 26 (72) | 10 (28) | 0.066 |
22 (61) | 14 (39) | 0.600 |
24 (67) | 12 (33) | 0.294 |
| Former | 6 (10) | 3 (50) | 3 (50) | 2 (33) | 4 (67) | 2 (33) | 4 (67) | |||
| Current | 19 (31) | 9 (47) | 10 (53) | 11 (58) | 8 (42) | 11 (58) | 8 (42) | |||
| Histology | ||||||||||
| ADC | 50 (82) | 35 (70) | 15 (30) | 0.014 |
34 (68) | 16 (32) | <0.001 |
35 (70) | 15 (30) | 0.002 |
| SCC | 5 (8) | 1 (20) | 4 (80) | 1 (20) | 4 (80) | 1 (20) | 4 (80) | |||
| Others | 6 (10) | 2 (33) | 4 (67) | 0 | 6 (100) | 1 (17) | 5 (83) | |||
| Tumor size (cm) | ||||||||||
| ≤ 3 | 26 (43) | 21 (81) | 5 (19) | 0.016 | 19 (73) | 7 (27) | 0.040 | 21 (81) | 5 (19) | 0.008 |
| > 3 | 35 (57) | 17 (49) | 18 (51) | 16 (46) | 19 (54) | 16 (46) | 19 (54) | |||
| Operation method | ||||||||||
| Biopsy | 1 (2) | 1 (100) | 0 | 1.000 | 1 (100) | 0 | 1.000 | 1 (100) | 0 | 1.000 |
| Resection | 60 (98) | 37 (62) | 23 (38) | 34 (57) | 26 (43) | 36 (60) | 24 (40) | |||
| EGFR TKI response | ||||||||||
| CR | 1 (2) | 1 (100) | 0 | 0.537 |
1 (100) | 0 | 0.122 |
1 (100) | 0 | 0.005 |
| PR | 13 (21) | 9 (69) | 4 (31) | 10 (77) | 3 (23) | 12 (92) | 1 (8) | |||
| SD | 14 (23) | 12 (86) | 2 (14) | 12 (86) | 2 (14) | 12 (86) | 2 (14) | |||
| PD | 33 (54) | 16 (48) | 17 (52) | 12 (36) | 21 (64) | 12 (36) | 21 (64) | |||
| n (%) | Objective response |
p-value | Disease control rate |
p-value | Median TTP (mo) | p-value | MST (mo) | p-value | |
|---|---|---|---|---|---|---|---|---|---|
| Total | 61 (100) | 14 (23) | 28 (46) | 16.0 | 30.0 | ||||
| Sex | |||||||||
| Female | 35 (57) | 7 (20) | 0.553 | 15 (43) | 0.613 | 18.0 | 0.440 | 29.0 | 0.445 |
| Male | 26 (43) | 7 (27) | 13 (50) | 13.5 | 35.0 | ||||
| Smoking history |
|||||||||
| Never | 36 (59) | 9 (25) | 0.762 |
17 (47) | 1.000 |
17.0 | 0.300 |
32.0 | 0.795 |
| Former | 6 (10) | 1 (17) | 3 (50) | 19.5 | 44.5 | ||||
| Current | 19 (31) | 4 (21) | 8 (42) | 11.0 | 28.0 | ||||
| Histology | |||||||||
| ADC | 50 (82) | 12 (24) | 0.726 |
26 (52) | 0.051 |
17.5 | 0.043 |
30.0 | 0.100 |
| SCC | 5 (8) | 1 (20) | 1 (20) | 11.0 | 28.0 | ||||
| Others | 6 (10) | 1 (17) | 1 (17) | 10.0 | 28.0 | ||||
| Direct sequencing | |||||||||
| EGFR mutation (+) | 38 (62) | 10 (26) | 0.537 | 22 (58) | 0.019 | 20.0 | 0.008 | 34.5 | 0.428 |
| EGFR mutation (-) | 23 (38) | 4 (17) | 6 (26) | 10.0 | 24.0 | ||||
| PNA clamping | |||||||||
| EGFR mutation (+) | 35 (57) | 11 (31) | 0.122 | 23 (66) | 0.001 | 19.0 | 0.020 | 34.0 | 0.606 |
| EGFR mutation (-) | 26 (43) | 3 (12) | 5 (23) | 10.5 | 24.0 | ||||
| Pyrosequencing | |||||||||
| EGFR mutation (+) | 37 (61) | 13 (35) | 0.005 | 25 (68) | <0.001 | 19.0 | 0.018 | 35.0 | 0.294 |
| EGFR mutation (-) | 24 (39) | 1 (4) | 3 (13) | 12.0 | 25.0 |
Never smokers were defined as patients who had a lifetime smoking exposure of <100 cigarettes and former smokers were defined as patients who had stopped smoking at least 1 yr before diagnosis; Comparison between never smokers and others; Comparison between adenocarcinoma and nonadenocarcinoma; Comparison between CR, PR and SD, PD.
Objective response: complete response or partial response; Disease control rate: complete response or partial response or stable disease; Never smokers were defined as patients who had a lifetime smoking exposure of <100 cigarettes and former smokers were defined as patients who had stopped smoking at least 1 yr before diagnosis; Comparison between never smokers and others; Comparison between adenocarcinoma and nonadenocarcinoma.

 E-submission
E-submission