Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 47(2); 2013 > Article
-
Original Article
Hedgehog Related Protein Expression in Breast Cancer: Gli-2 Is Associated with Poor Overall Survival - Soyoung Im, Hyun Joo Choi, Changyoung Yoo, Ji-Han Jung, Ye-Won Jeon1, Young Jin Suh1, Chang Suk Kang2
-
Korean Journal of Pathology 2013;47(2):116-123.
DOI: https://doi.org/10.4132/KoreanJPathol.2013.47.2.116
Published online: April 24, 2013
Department of Hospital Pathology, St. Vincent's Hospital, The Catholic University of Korea College of Medicine, Suwon, Korea.
1Department of Surgery, St. Vincent's Hospital, The Catholic University of Korea College of Medicine, Suwon, Korea.
2Department of Hospital Pathology, St. Mary's Hospital, The Catholic University of Korea College of Medicine, Seoul, Korea.
- Corresponding Author: Hyun Joo Choi, M.D. Department of Pathology, St. Vincent's Hospital, The Catholic University of Korea College of Medicine, 93 Jungbu-daero, Paldal-gu, Suwon 442-723, Korea. Tel: +82-31-249-7592, Fax: +82-31-244-6786, chj0103@catholic.ac.kr
© 2013 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- The hedgehog (Hh) signaling pathway is known to play a critical role in various malignancies, but its clinicopathologic role in breast cancer is yet to be established.
-
Methods
- Tissue microarray blocks from 334 cases of breast cancer were prepared. The expression of six Hh signaling proteins including sonic hedgehog (Shh), patched (Ptch), smoothened (Smo), and the glioma-associated oncogene (Gli)-1, Gli-2, and Gli-3 were analyzed immunohistochemically.
-
Results
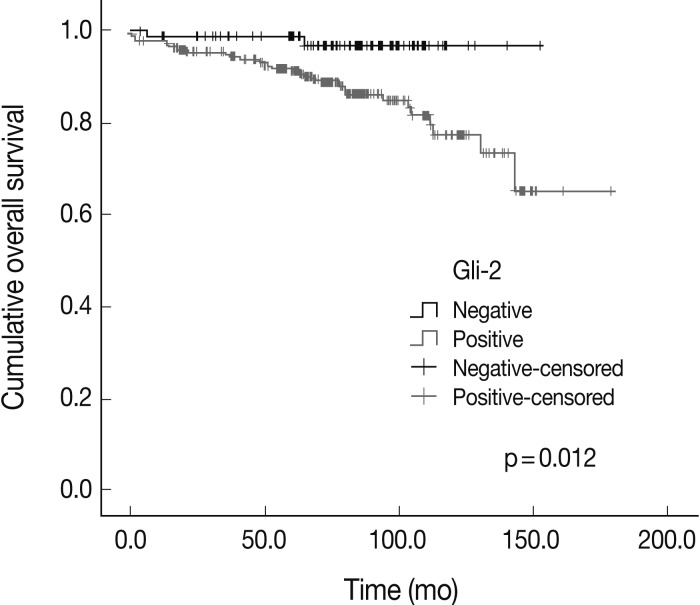
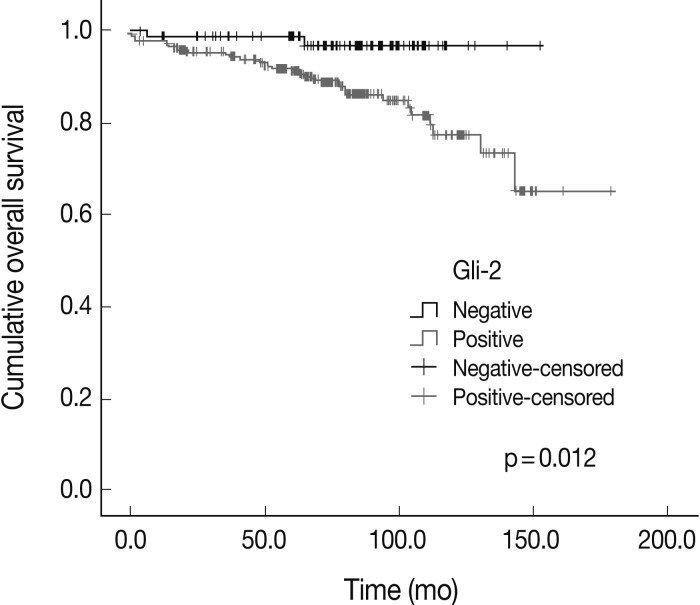
- The expression of Hh signaling proteins was significantly correlated with some prognostic factors including the correlation of lymph node metastasis with the expression of Shh (p=0.001) and Ptch (p=0.064), the correlation of the stages with Shh and Gli-3 expression (p=0.007 and p=0.024, respectively), the correlation of the nuclear grade with the Smo (p=0.004) and Gli-3 (p=0.000), and the correlation of the histologic grade with the Ptch (p=0.016), Smo (p=0.007), and Gli-3 (p=0.000). The Shh, Ptch, Smo, Gli-1, and Gli-2 expression was significantly different between the phenotypes (p=0.000, p=0.001, p=0.004, p=0.039, and p=0.031, respectively). Gli-2 expression was correlated with a worse overall survival outcome (p=0.012).
-
Conclusions
- Hh pathway activation is correlated with a more aggressive clinical behavior in breast carcinomas. The comparison of phenotypes suggested that the Hh pathway may be a useful therapeutic target for breast carcinoma. Patients with Gli-2 expression had a significantly lower overall survival rate and, therefore, it showed promise as a prognostic marker.
- Clinical samples
- A total of 334 patients with primary breast carcinoma underwent resection at St. Vincent's Hospital (Suwon, Korea) between January 1999 and August 2007. Formalin fixed paraffin embedded (FFPE) tissue and clinical data were collected retrospectively. All of the archival hematoxylin and eosin (H&E) stained slides for each patient were reviewed. The nuclear grades and histologic grades were evaluated according to the Nottingham grading system.9 The tumor stages were evaluated according to the 7th edition of the American Joint Committee on Cancer (AJCC) criteria. A total of 322 cases were available with patient survival data. This study was approved by the Institutional Review Board of The Catholic University of Korea (VC10SISI0093).
- Tissue microarray (TMA)
- TMA blocks were constructed containing 334 invasive carcinoma specimens from the FFPE tissue. The representative tumor site was chosen on H&E slides, and the site corresponding to the confirmed tumor site in the paraffin block was marked. The selected area was harvested using a 2 mm Quick-Ray tip-punch (Micro Digital Co., Seoul, Korea) and was transferred to a TMA mold. The H&E stained slides of the TMA blocks confirmed whether or not the appropriate tumor site had been selected.
- Immunohistochemistry
- Immunohistochemical staining was conducted on 5 µm sections of the TMA blocks. The paraffin sections were mounted on poly-L-lysin coated glass slides, deparaffinized, and then they were rehydrated in a graded series of ethanol, followed by microwave antigen retrieval. Endogenous peroxidase activity was blocked by treating the slides with 3% H2O2 in methanol for 10 minutes at room temperature. The primary antibodies were incubated overnight at 4℃. The following primary antibodies were used: Shh (1:200, EP1190Y, Epitomics, Burlingame, CA, USA), Ptch (1:200, ab53715, Abcam, Cambridge, MA, USA), Smo (1:100, ab72130, Abcam), Gli-1 (1:50, ab92611, Abcam), Gli-2 (1:400, ab7181, Abcam), Gli-3 (1:400, PAB-10052, Abnova, Taipei, Taiwan), ER (1:300, 6F11, Novocastra, Newcastle upon Tyne, UK), progesterone receptor (PR; 1:600, 16, Novocastra), HER2 (1:1,800, polyclonal, Dako, Glostrup, Denmark), and Ki-67 (1:200, MM1, Novocastra). Immunostaining was conducted using the rabbit or mouse DAKO ChemMate EnVision system and Peroxidase/DAB kit (Dako). The sections were then counterstained with Meyer hematoxylin and then dehydrated, cleared, and mounted.
- The immunohistochemical staining and the histology were examined independently by two pathologists. The cases with discrepant scores were discussed in order to obtain a consensus. The immunoreactivity was interpreted in the cytoplasm for the Shh, Ptch, and Smo, and in the nucleus for the Gli-1, Gli-2, and Gli-3. The staining intensity was graded as follows: 0, negative; 1, weak; 2, moderate; and 3, intense; and the proportion was graded according to the percentage of positive cells as follows: 0, <5%; 1, 5-25%; 2, 26-50%; 3, 51-75%; and 4, >75%. The intensity score and proportion score were multiplied in order to generate an immunoreactive score (IS). Negative expression was defined as an IS lower than 4.
- Tumor phenotype classification
- The breast cancer phenotypes were classified according to the 2011 St. Gallen consensus.10 The immunohistochemical staining for the ER and PR was evaluated using the Allred method.11 An Allred score of 3 or higher was considered positive. The immmunohistochemical results for HER2 were defined as follows: 0, no staining; 1+, weak incomplete membranous staining in any proportion of tumor cells; 2+, complete membranous staining weak in intensity of >10% of tumor cells; and 3+, intense complete membrane staining of >30% of the invasive tumor cells. Values of 0 and 1+ were considered negative; 2+, equivocal; and 3+, positive. The equivocal cases were reassigned after silver in situ hybridization. The phenotypes were classified as follows; luminal A (LumA) type: ER and/or PR positive, HER2 negative, and Ki-67 index<14%; luminal B HER2 negative (LumB HER2 neg) type: ER and/or PR positive, HER2 negative, and Ki-67 index≥15%; luminal B HER2 positive (LumB HER2 pos) type: ER and/or PR positive, HER2 positive, and any Ki-67 index; HER2 positive (HER2) type: ER, PR negative, and HER2 positive; and triple negative type: ER, PR, and HER2 negative.
- Statistical analysis
- Statistical analyses were performed by using SPSS for Windows ver. 13.0 (SPSS Inc., Chicago, IL, USA). The two-sided p-values were determined via chi-square tests. The patients' overall survival was analyzed using the Kaplan-Meier method with the use of the log-rank test for univariate analysis. A value of p<0.05 was considered significant for all of the analyses.
MATERIALS AND METHODS
- Patient characteristics
- The mean age of the 334 total patients was 51.3 years (range, 30 to 82 years). The clinicopathological data of our study population are presented in Table 1. Follow-up data were available for 322 patients and the median follow-up duration was 75.1 months (range, 0 to 179 months) after the primary surgery. Twenty-one (6.5%) patients had local recurrence, 62 (19.3%) patients developed distant metastasis and 31 (9.6%) patients died during follow up.
- Immunohistochemical results for Shh, Ptch, Smo, Gli-1, Gli-2, and Gli-3
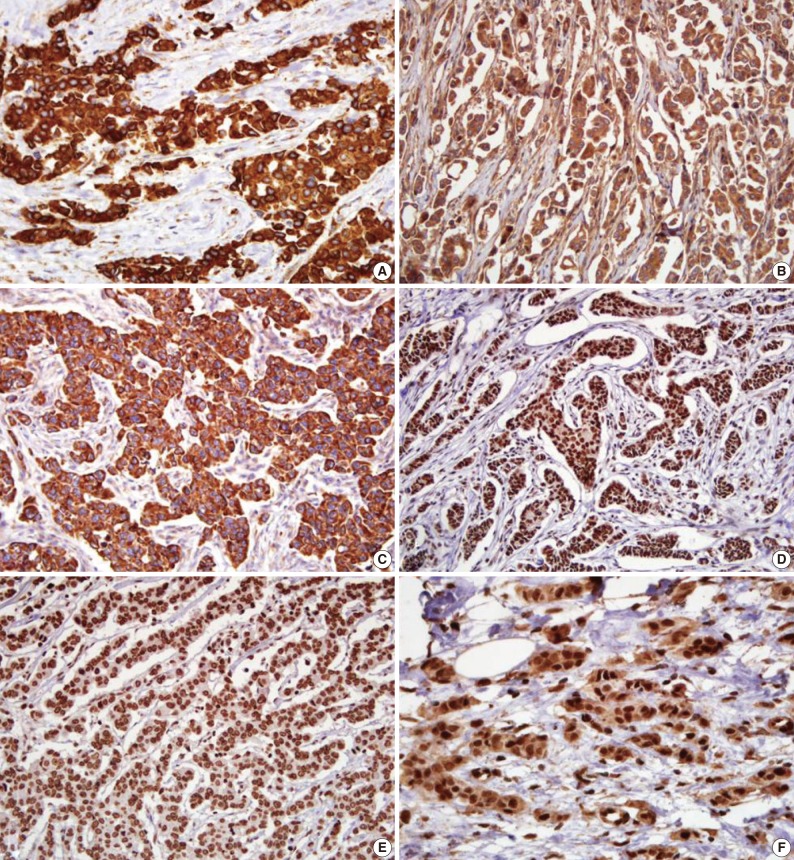
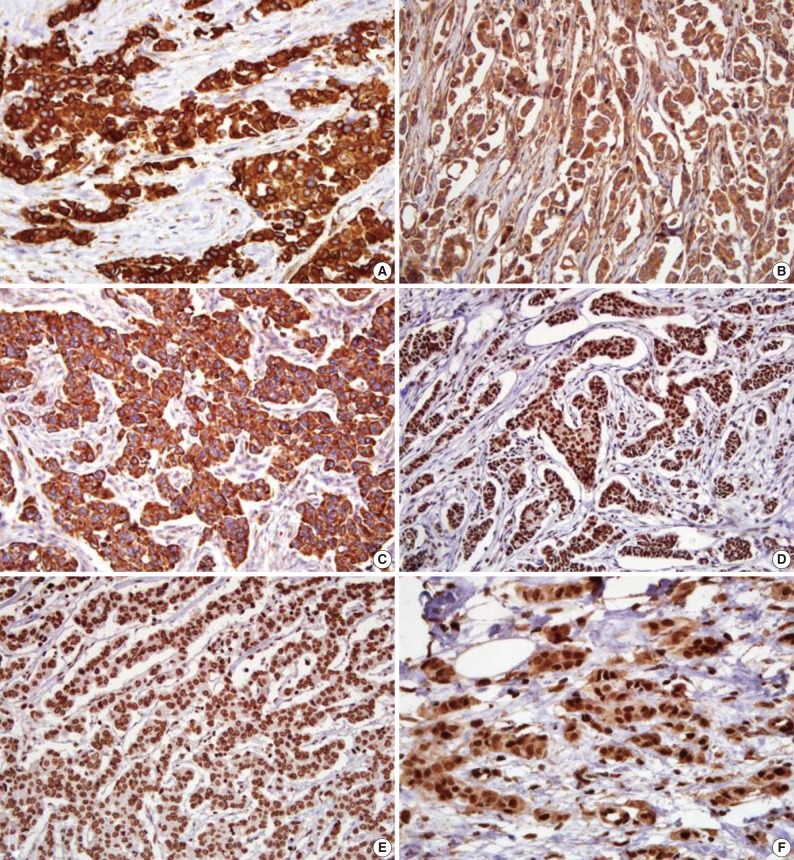
- The microscopic features of the immunohistochemical staining for each protein are presented in Fig. 1. Shh, Ptch, and Smo were stained in the cytoplasm. Gli-1, Gli-2, and Gli-3 had a nuclear staining pattern. Of the 334 breast carcinoma cases, the Shh, Ptch, Smo, Gli-1, Gli-2, and Gli-3 were expressed in 164 (49.1%), 70 (21.0%), 75 (22.5%), 141 (42.2%), 252 (75.4%), and 37 (11.1%) cases, respectively.
- Relationship between the expression of Shh, Ptch, Smo, Gli-1, Gli-2, and Gli-3 and the clinicopathological factors
- The correlations between the Shh, Ptch, Smo, Gli-1, Gli-2, and Gli-3, and the clinicopathological variables are presented in Table 1. Lymph node metastasis was correlated with the expression of Shh (p=0.001) and Ptch (p=0.064). Shh and Gli-3 expression was significantly correlated with the stage (p=0.007 and p=0.024, respectively). The nuclear grade was related to the Smo (p=0.004) and Gli-3 (p=0.000). The histologic grade was related to the Ptch (p=0.016), Smo (p=0.007), and Gli-3 (p=0.000). The ER expression was related to the Ptch (p=0.012), Smo (p=0.000), Gli-1 (p=0.008), and Gli-2 (p=0.000). The PR expression was related to the Smo (p=0.001) and Gli-1 (p=0.031). HER2 showed a significant correlation with the Shh (p=0.000) and Gli-3 (p=0.002).
- Relationship between the Hh related protein expression and the breast cancer phenotype
- The correlation between the Shh, Ptch, Smo, Gli-1, Gli-2, and Gli-3, and the breast cancer phenotypes according to the 2011 St. Gallen consensus are presented in Table 2. Shh, Ptch, Smo, Gli-1, and Gli-2 expression varied significantly between phenotypes (p=0.000, p=0.001, p=0.004, p=0.039, and p=0.031, respectively). Shh expression was observed in 143 (49.3%) cases and was higher in the HER2 type (74.1%). Ptch expression was present in 70 (21.0%) cases, and was higher in the LumB HER2 neg type (44.2%). Smo expression was observed in 65 (22.4%) cases and was higher in the HER2 type and triple negative type (31.0% and 30.9%, respectively). Gli-1 expression was present in 119 (41.0%) cases and was higher in the LumB HER2 pos type (64.0%). Gli-2 expression was observed in 215 (74.1%) cases and was higher in the luminal types: LumA (85.1%), LumB HER2 neg (81.4%), and LumB HER2 pos (80.0%). Gli-3 expression did not differ significantly among the phenotypes (p=0.282).
- Survival analysis
- The overall survival rates according to the clinicopathologic and immunohistochemical parameters are summarized in Table 3. The stage (p=0.000) was significantly correlated with the patients' overall survival. Among the Hh related proteins that were analyzed, the Gli-2 positive group had a significantly worse survival rate than the Gli-2 negative group (p=0.012) (Fig. 2). The Shh, Ptch, Smo, Gli-1, and Gli-3 did not show statistical significance related to the patients' prognosis.
RESULTS
- The Hh signaling pathway, initially discovered in the Drosophila, is known to play a crucial role in controlling cell fate and patterning during embryogenesis.12 Disruption of the Hh signaling pathway results in congenital brain, facial, and other midline defects. Mutational activation of the Hh signaling pathway is primarily associated with basal cell carcinoma and medulloblastoma.3,4 Ligand-dependent activation has also been reported in various types of malignancies, such as small cell lung carcinoma and carcinomas of the stomach, pancreas, prostate, uterine cervix, colon and breasts.2,6,7,13-20
- Kubo et al.2 evaluated the expression of Shh, Ptch, and Gli-1 by immunohistochemical staining in 52 cases of human breast carcinoma. Tumor cells overexpressed the Shh, Ptch and Gli-1 in 52, 50, and 52 of the 52 total specimens, respectively. In addition, four human breast carcinoma cell lines showed increased expression of Shh, Ptch, and Gli-1. However, according to their results, there was not a statistically significant difference between the T stage, lymph node involvement, and overall tumor node metastasis (TNM) stage. Ten Haaf et al.21 determined that elevated levels of the Gli-1 protein in human breast cancer were correlated with aggressive behavior of the cancer, such as higher tumor stages and lymph node status. Our data suggests that the overexpression of Hh related proteins are associated with a larger tumor size (Gli-3), higher lymph node status (Shh, Ptch, and Gli-3), advanced stages (Shh and Ptch), higher nuclear grades (Smo and Gli-3), and higher histological grades (Smo), all of which indicate more aggressive tumor behavior.
- There has been limited data on Hh related proteins and the actual overall survival in patients with breast carcinomas. According to ten Haaf et al.,21 the high Gli-1 expression groups had an unfavorable overall survival rate. O'Toole et al.18 evaluated Hh overexpression in a group of 279 patients with invasive ductal carcinoma of the breast, and reported that the expression of the Hh ligand was significantly associated with an increased risk of metastasis and breast-cancer specific death. There are a few studies on Hh related protein expression and the survival of malignancies in other organs. Zhang et al.22 reported that Gli-2 expression was correlated with poor prognosis in patients with hepatocellular carcinomas. Ding et al.23 reported that post-operative liver metasis-free survival was significantly lower with high Smo expression in colon cancer. On the other hand, Lee et al.24 reported that the Ptch and Gli-2 positive groups had a better overall survival rate for gastric adenocarcinomas. However, Gli-2 expression showed a worse overall survival rate in breast carcinomas in our study, which supported the previously discussed results that Hh related protein expression is correlated with clinicopathologic features resulting in more aggressive behavior.
- Gli-2, one of three members of the mammalian Gli transcription factor family, is primarily known as a transcriptional activator that mediates the cellular response of Hh signaling.3 Lewis25 demonstrated that disruption of Gli-2 in mouse mammary gland development led to ductal dysplasias that closely resembled some hyperplasia of human breast. However, there is only limited data available regarding Gli-2 and human breast cancer. According to Xuan and Lin,26 Gli-2 expression is related to PR expression, the Ki-67 proliferating index, node metastasis, and the clinical stage. Our study is the first to report that Gli-2 expression had a worse overall survival rate in breast carcinomas. This result suggests the possibility that Gli-2 could be used as a prognostic marker to select patients for aggressive treatment.
- In 2011, the 12th St. Gallen International Breast Cancer Conference Expert Panel adopted a new approach to the classification of patients for therapeutic purposes.10 For practical reasons, this approach uses immunohistochemical results of ER and PR, the HER2 status, and the Ki-67 proliferating index in order to identify tumor subtypes. The additional Ki-67 proliferating index has divided the previously known 'luminal A' type (ER and/or PR positive, HER2 negative) into LumA (ER and/or PR positive, HER2 negative, and Ki-67 index<14%) and LumB HER2 neg type (ER and/or PR positive, HER2 negative, and Ki-67 index≥15%). The key to this new classification is to coordinate the subtypes with treatment. The LumB HER2 neg type may need systemic chemotherapy due to the high Ki-67 index.
- According to our results, Shh expression was higher in the HER2 type, Ptch expression was higher in the LumB HER2 neg type, and Gli-1 expression was higher in the LumB HER2 pos type. These phenotypes all need additional cytotoxic chemotherapy in addition to endocrine therapy or trastuzumab. The Hh pathway could be a therapeutic target for chemotherapy in these cases. The Smo expression was significantly higher in both the HER2 and triple negative types, which also require cytotoxic agents for chemotherapy. The steroid alkaloid cyclopamine inhibits the Hh pathway activation via the direct bonding of Smo.27 This effect suggests that cyclopamine derivatives could be a possible therapeutic agents for breast carcinomas. The Gli-2 expression was higher in the LumA, LumB HER2 neg, and LumB HER2 pos types. These three types are ER and/or PR positive types. Xuan and Lin26 reported that Gli-2 expression was related to PR expression. However, the full relationship between Gli-2 and hormone receptors has yet to be discovered.
- In summary, our data showed that the expression of Shh, Ptch, Smo, Gli-1, Gli-2, and Gli-3 was significantly correlated with a variety of clinicopathological indicators, indicating that Hh pathway activation is correlated with more aggressive clinical behavior. Most of all, patients with Gli-2 expression had a considerably worse prognosis based on overall survival rates, which showed that Gli-2 is a promising prognostic marker. Evaluation of the Hh related proteins may predict a patient's outcome and be useful in selecting patients for aggressive treatment. The Hh related protein expression comparison between phenotypes suggests that the Hh pathway may be a useful therapeutic target for chemotherapy in breast carcinomas.
DISCUSSION
Acknowledgments
Acknowledgments
- 1. Toftgård R. Hedgehog signalling in cancer. Cell Mol Life Sci 2000; 57: 1720-1731. ArticlePubMedPMCPDF
- 2. Kubo M, Nakamura M, Tasaki A, et al. Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer. Cancer Res 2004; 64: 6071-6074. ArticlePubMedPDF
- 3. Gupta S, Takebe N, Lorusso P. Targeting the hedgehog pathway in cancer. Ther Adv Med Oncol 2010; 2: 237-250. ArticlePubMedPMCPDF
- 4. Katano M. Hedgehog signaling pathway as a therapeutic target in breast cancer. Cancer Lett 2005; 227: 99-104. ArticlePubMed
- 5. Karlstrom RO, Tyurina OV, Kawakami A, et al. Genetic analysis of zebrafish gli1 and gli2 reveals divergent requirements for gli genes in vertebrate development. Development 2003; 130: 1549-1564. ArticlePubMedPDF
- 6. Xuan YH, Jung HS, Choi YL, et al. Enhanced expression of hedgehog signaling molecules in squamous cell carcinoma of uterine cervix and its precursor lesions. Mod Pathol 2006; 19: 1139-1147. ArticlePubMedPDF
- 7. Onishi H, Katano M. Hedgehog signaling pathway as a therapeutic target in various types of cancer. Cancer Sci 2011; 102: 1756-1760. ArticlePubMed
- 8. Koga K, Nakamura M, Nakashima H, et al. Novel link between estrogen receptor alpha and hedgehog pathway in breast cancer. Anticancer Res 2008; 28: 731-740. PubMed
- 9. Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 1991; 19: 403-410. ArticlePubMed
- 10. Goldhirsch A, Wood WC, Coates AS, et al. Strategies for subtypes: dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol 2011; 22: 1736-1747. ArticlePubMedPMC
- 11. Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol 1998; 11: 155-168. PubMed
- 12. Cohen MM Jr. The hedgehog signaling network. Am J Med Genet A 2003; 123A: 5-28. ArticlePubMed
- 13. Ma X, Chen K, Huang S, et al. Frequent activation of the hedgehog pathway in advanced gastric adenocarcinomas. Carcinogenesis 2005; 26: 1698-1705. ArticlePubMed
- 14. Bian YH, Huang SH, Yang L, Ma XL, Xie JW, Zhang HW. Sonic hedgehog-Gli1 pathway in colorectal adenocarcinomas. World J Gastroenterol 2007; 13: 1659-1665. ArticlePubMedPMC
- 15. Barginear MF, Leung M, Budman DR. The hedgehog pathway as a therapeutic target for treatment of breast cancer. Breast Cancer Res Treat 2009; 116: 239-246. ArticlePubMedPDF
- 16. Kameda C, Tanaka H, Yamasaki A, et al. The hedgehog pathway is a possible therapeutic target for patients with estrogen receptor-negative breast cancer. Anticancer Res 2009; 29: 871-879. PubMed
- 17. O'Toole SA, Swarbrick A, Sutherland RL. The hedgehog signalling pathway as a therapeutic target in early breast cancer development. Expert Opin Ther Targets 2009; 13: 1095-1103. ArticlePubMed
- 18. O'Toole SA, Machalek DA, Shearer RF, et al. Hedgehog overexpression is associated with stromal interactions and predicts for poor outcome in breast cancer. Cancer Res 2011; 71: 4002-4014. ArticlePubMedPDF
- 19. Das S, Samant RS, Shevde LA. The hedgehog pathway conditions the bone microenvironment for osteolytic metastasis of breast cancer. Int J Breast Cancer 2012; 2012: 298623.ArticlePubMedPDF
- 20. Fiaschi M, Rozell B, Bergström A, Toftgård R. Development of mammary tumors by conditional expression of GLI1. Cancer Res 2009; 69: 4810-4817. ArticlePubMedPMCPDF
- 21. ten Haaf A, Bektas N, von Serenyi S, et al. Expression of the glioma-associated oncogene homolog (GLI) 1 in human breast cancer is associated with unfavourable overall survival. BMC Cancer 2009; 9: 298.PubMedPMCPDF
- 22. Zhang D, Cao L, Li Y, Lu H, Yang X, Xue P. Expression of glioma-associated oncogene 2 (Gli 2) is correlated with poor prognosis in patients with hepatocellular carcinoma undergoing hepatectomy. World J Surg Oncol 2013; 11: 25.ArticlePubMedPMCPDF
- 23. Ding YL, Wang QS, Zhao WM, Xiang L. Expression of smoothened protein in colon cancer and its prognostic value for postoperative liver metastasis. Asian Pac J Cancer Prev 2012; 13: 4001-4005. ArticlePubMed
- 24. Lee SJ, Do IG, Lee J, et al. Gastric cancer (GC) patients with hedgehog pathway activation: PTCH1 and GLI2 as independent prognostic factors. Target Oncol 2013 2 01 [Epub]. http://dx.doi.org/10.1007/s11523-013-0253-1. ArticlePDF
- 25. Lewis MT. Hedgehog signaling in mouse mammary gland development and neoplasia. J Mammary Gland Biol Neoplasia 2001; 6: 53-66. ArticlePubMedPDF
- 26. Xuan Y, Lin Z. Expression of Indian hedgehog signaling molecules in breast cancer. J Cancer Res Clin Oncol 2009; 135: 235-240. ArticlePubMedPDF
- 27. Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of hedgehog signaling by direct binding of cyclopamine to smoothened. Genes Dev 2002; 16: 2743-2748. ArticlePubMedPMC
REFERENCES
| Clinicopathologic factors | No. of cases (%) |
No. of positive cases (%) |
|||||
|---|---|---|---|---|---|---|---|
| Shh | Ptch | Smo | Gli-1 | Gli-2 | Gli-3 | ||
| Age (yr) | |||||||
| < 50 | 179 (53.6) | 87 (48.6) | 100 (55.9) | 47 (26.3) | 77 (43.0) | 141 (78.8) | 99 (55.3) |
| > 50 | 155 (46.4) | 77 (49.7) | 83 (53.5) | 28 (18.1) | 64 (41.3) | 111 (71.6) | 92 (59.4) |
| p-value | 0.845 | 0.671 | 0.074 | 0.750 | 0.130 | 0.456 | |
| Size (cm) | |||||||
| < 2 | 129 (38.6) | 60 (46.5) | 72 (55.8) | 30 (23.3) | 57 (44.2) | 95 (73.6) | 85 (65.9) |
| 2-5 | 172 (51.5) | 84 (48.8) | 91 (52.9) | 38 (22.1) | 71 (41.3) | 129 (75.0) | 93 (54.1) |
| > 5 | 33 (9.9) | 20 (60.6) | 20 (60.6) | 7 (21.2) | 13 (39.4) | 28 (84.8) | 13 (39.4) |
| p-value | 0.350 | 0.687 | 0.956 | 0.829 | 0.403 | 0.011* | |
| Node metastasis | |||||||
| 0 | 179 (53.6) | 84 (46.9) | 96 (53.6) | 40 (22.3) | 80 (44.7) | 136 (76.0) | 111 (62.0) |
| 1-3 | 98 (29.3) | 40 (40.8) | 47 (48.0) | 18 (18.4) | 36 (36.7) | 74 (75.5) | 36 (36.7) |
| > 4 | 57 (17.1) | 40 (70.2) | 40 (70.2) | 17 (29.8) | 25 (43.9) | 42 (73.7) | 44 (77.2) |
| p-value | 0.001* | 0.025* | 0.257 | 0.423 | 0.940 | 0.000* | |
| Stage | |||||||
| I | 78 (23.4) | 32 (41.0) | 45 (57.7) | 18 (23.1) | 36 (46.2) | 57 (73.1) | 54 (69.2) |
| II | 169 (50.6) | 78 (46.2) | 83 (49.1) | 36 (21.3) | 67 (39.6) | 126 (74.6) | 89 (52.7) |
| III | 81 (24.3) | 48 (59.3) | 49 (60.5) | 19 (23.5) | 33 (40.7) | 63 (77.8) | 45 (55.6) |
| IV | 6 (1.8) | 6 (100.0) | 6 (100.0) | 2 (33.3) | 5 (83.3) | 6 (100.0) | 3 (50.0) |
| p-value | 0.007* | 0.037* | 0.896 | 0.159 | 0.475 | 0.100 | |
| Nuclear grade | |||||||
| 1 | 39 (11.7) | 20 (51.3) | 23 (59.0) | 4 (10.3) | 15 (38.5) | 34 (87.2) | 18 (46.2) |
| 2 | 168 (50.3) | 79 (47.0) | 90 (53.6) | 31 (18.5) | 68 (40.5) | 128 (76.2) | 89 (53.0) |
| 3 | 127 (38.0) | 65 (51.2) | 70 (55.1) | 40 (31.5) | 58 (45.7) | 90 (70.9) | 84 (66.1) |
| p-value | 0.747 | 0.826 | 0.004* | 0.590 | 0.112 | 0.026* | |
| Histologic grade | |||||||
| 1 | 75 (22.5) | 34 (45.3) | 40 (53.3) | 9 (12.0) | 35 (46.7) | 61 (81.3) | 42 (56.0) |
| 2 | 136 (40.7) | 64 (47.1) | 73 (53.7) | 28 (20.6) | 61 (44.9) | 105 (77.2) | 81 (59.6) |
| 3 | 123 (36.8) | 66 (53.7) | 70 (56.9) | 38 (30.9) | 45 (36.6) | 86 (69.9) | 68 (55.3) |
| p-value | 0.433 | 0.837 | 0.007* | 0.273 | 0.160 | 0.764 | |
| ER | |||||||
| – | 154 (46.1) | 79 (51.3) | 67 (43.5) | 48 (31.2) | 53 (34.4) | 102 (66.2) | 75 (48.7) |
| + | 180 (53.9) | 85 (47.2) | 116 (64.4) | 27 (15.0) | 88 (48.9) | 150 (83.3) | 116 (64.4) |
| p-value | 0.458 | 0.000* | 0.000* | 0.008* | 0.000* | 0.004* | |
| PR | |||||||
| – | 184 (55.1) | 95 (51.6) | 97 (52.7) | 54 (29.3) | 68 (37.0) | 139 (75.5) | 92 (50.0) |
| + | 150 (44.9) | 69 (46.0) | 86 (57.3) | 21 (14.0) | 73 (48.7) | 113 (75.3) | 99 (66.0) |
| p-value | 0.306 | 0.399 | 0.001* | 0.031* | 0.965 | 0.003* | |
| HER2 | |||||||
| – | 251 (75.1) | 108 (43.0) | 144 (57.4) | 52 (20.7) | 107 (42.6) | 193 (76.9) | 144 (57.4) |
| + | 83 (24.9) | 56 (67.5) | 39 (47.0) | 23 (27.7) | 34 (41.0) | 59 (71.1) | 47 (56.6) |
| p-value | 0.000* | 0.099 | 0.186 | 0.790 | 0.287 | 0.905 | |
| Total (n = 290)a | LumA (n = 67) | LumB HER2 neg (n = 43) | LumB HER2 pos (n = 25) | HER2 (n = 58) | Triple negative (n = 97) | p-value | |
|---|---|---|---|---|---|---|---|
| Shh | |||||||
| – | 147 (50.7) | 33 (49.3) | 27 (62.8) | 12 (48.0) | 15 (25.9) | 60 (61.9) | 0.000* |
| + | 143 (49.3) | 34 (50.7) | 16 (37.2) | 13 (52.0) | 43 (74.1) | 37 (38.1) | |
| Ptch | |||||||
| – | 132 (45.5) | 21 (31.3) | 10 (23.3) | 13 (52.0) | 31 (53.4) | 57 (58.8) | 0.000* |
| + | 158 (54.5) | 46 (68.7) | 33 (76.7) | 12 (48.0) | 27 (46.6) | 40 (41.2) | |
| Smo | |||||||
| – | 225 (77.6) | 58 (86.6) | 40 (93.0) | 20 (80.0) | 40 (69.0) | 67 (69.1) | 0.004* |
| + | 65 (22.4) | 9 (13.4) | 3 (7.0) | 5 (20.0) | 18 (31.0) | 30 (30.9) | |
| Gli-1 | |||||||
| – | 171 (59.0) | 35 (52.2) | 25 (58.1) | 9 (36.0) | 40 (69.0) | 62 (63.9) | 0.039* |
| + | 119 (41.0) | 32 (47.8) | 18 (41.9) | 16 (64.0) | 18 (31.0) | 35 (36.1) | |
| Gli-2 | |||||||
| – | 75 (25.9) | 10 (14.9) | 8 (18.6) | 5 (20.0) | 20 (34.5) | 32 (33.0) | 0.031* |
| + | 215 (74.1) | 57 (85.1) | 35 (81.4) | 20 (80.0) | 38 (65.5) | 65 (67.0) | |
| Gli-3 | |||||||
| – | 122 (42.1) | 17 (25.4) | 20 (46.5) | 6 (24.0) | 30 (51.7) | 49 (50.5) | 0.003* |
| + | 168 (57.9) | 50 (74.6) | 23 (53.5) | 19 (76.0) | 28 (48.3) | 48 (49.5) |
Values are presented as number (%).
LumA, luminal A; LumB HER2 neg, luminal B HER2 negative; LumB HER pos, luminal B HER2 positive; Shh, sonic hedgehog; Ptch, patched; Smo, smoothened; Gli, glioma-associated oncogenes.
*Statistically significant (p<0.05).
aClassification is only available in 290 cases.
|
Breast carcinoma (n = 322) |
||||
|---|---|---|---|---|
| Total | No. of deceased | p-value | ||
| Age (yr) | < 50 | 170 | 17 | 0.233 |
| ≥ 50 | 152 | 19 | ||
| Stage | I | 73 | 2 | 0.000* |
| II | 163 | 14 | ||
| III | 80 | 16 | ||
| IV | 6 | 4 | ||
| Phenotype | LumA | 67 | 1 | 0.130 |
| LumB HER2 neg | 43 | 8 | ||
| LumB HER2 pos | 25 | 1 | ||
| HER2 | 58 | 5 | ||
| Triple negative | 97 | 9 | ||
| Shh | – | 167 | 17 | 0.447 |
| + | 155 | 19 | ||
| Ptch | – | 147 | 15 | 0.638 |
| + | 175 | 21 | ||
| Smo | – | 252 | 27 | 0.695 |
| + | 70 | 9 | ||
| Gli-1 | – | 186 | 25 | 0.354 |
| + | 136 | 11 | ||
| Gli-2 | – | 79 | 2 | 0.012* |
| + | 243 | 34 | ||
| Gli-3 | – | 139 | 21 | 0.469 |
| + | 183 | 15 | ||
Figure & Data
References
Citations

- Dysregulation of deubiquitination in breast cancer
Lili Kong, Xiaofeng Jin
Gene.2024; 902: 148175. CrossRef - siRNA treatment targeting integrin α11 overexpressed via EZH2-driven axis inhibits drug-resistant breast cancer progression
Prakash Chaudhary, Kiran Yadav, Ho Jin Lee, Keon Wook Kang, Jongseo Mo, Jung-Ae Kim
Breast Cancer Research.2024;[Epub] CrossRef - Implicating clinical utility of altered expression of PTCH1 & SMO in oral squamous cell carcinoma
Hitarth V. Patel, Jigna S. Joshi, Franky D. Shah
Journal of Molecular Histology.2024; 55(4): 379. CrossRef - A clinicopathological exploration of Hedgehog signaling: implications in oral carcinogenesis
Hitarth V. Patel, Jigna S. Joshi, Franky D. Shah
Journal of Cancer Research and Clinical Oncology.2023; 149(18): 16525. CrossRef - GLI3 and androgen receptor are mutually dependent for their malignancy-promoting activity in ovarian and breast cancer cells
Min Lin, Haiyan Zhu, Qi Shen, Lu-Zhe Sun, Xueqiong Zhu
Cellular Signalling.2022; 92: 110278. CrossRef - Persistent Properties of a Subpopulation of Cancer Cells Overexpressing the Hedgehog Receptor Patched
Álvaro Javier Feliz Morel, Anida Hasanovic, Aurélie Morin, Chloé Prunier, Virginie Magnone, Kevin Lebrigand, Amaury Aouad, Sarah Cogoluegnes, Judith Favier, Claude Pasquier, Isabelle Mus-Veteau
Pharmaceutics.2022; 14(5): 988. CrossRef - Case Report: Submucosal gastroblastoma with a novel PTCH1::GLI2 gene fusion in a 58-year-old man
Cuimin Chen, Junliang Lu, Huanwen Wu
Frontiers in Oncology.2022;[Epub] CrossRef - Higher Expressions of SHH and AR Are Associated with a Positive Receptor Status and Have Impact on Survival in a Cohort of Croatian Breast Cancer Patients
Ivan Budimir, Čedna Tomasović-Lončarić, Kristina Kralik, Josipa Čonkaš, Domagoj Eljuga, Rado Žic, Božo Gorjanc, Hrvoje Tucaković, Doroteja Caktaš, Josip Jaman, Valentino Lisek, Zlatko Vlajčić, Krešimir Martić, Petar Ozretić
Life.2022; 12(10): 1559. CrossRef - New insight into the role of PTCH1 protein in serous ovarian carcinomas
Valentina Karin‑Kujundzic, Adriana Covarrubias‑Pinto, Anita Skrtic, Semir Vranic, Ljiljana Serman
International Journal of Oncology.2022;[Epub] CrossRef - Hedgehog gene expression patterns among intrinsic subtypes of breast cancer: Prognostic relevance
Araceli García-Martínez, Ariadna Pérez-Balaguer, Fernando Ortiz-Martínez, Eloy Pomares-Navarro, Elena Sanmartín, Marta García-Escolano, Yoel G. Montoyo-Pujol, Elena Castellón-Molla, Gloria Peiró
Pathology - Research and Practice.2021; 223: 153478. CrossRef - Coexpression of Epha10 and Gli3 Promotes Breast Cancer Cell Proliferation, Invasion and Migration
Jing Peng, Danhua Zhang
Journal of Investigative Medicine.2021; 69(6): 1215. CrossRef - The Role of Smoothened-Dependent and -Independent Hedgehog Signaling Pathway in Tumorigenesis
Jian Yi Chai, Vaisnevee Sugumar, Mohammed Abdullah Alshawsh, Won Fen Wong, Aditya Arya, Pei Pei Chong, Chung Yeng Looi
Biomedicines.2021; 9(9): 1188. CrossRef - HER2-mediated GLI2 stabilization promotes anoikis resistance and metastasis of breast cancer cells
Parul Gupta, Nehal Gupta, Neel M. Fofaria, Alok Ranjan, Sanjay K. Srivastava
Cancer Letters.2019; 442: 68. CrossRef - Role of Hedgehog Signaling in Breast Cancer: Pathogenesis and Therapeutics
Natalia Riobo-Del Galdo, Ángela Lara Montero, Eva Wertheimer
Cells.2019; 8(4): 375. CrossRef - Carbonic anhydrase XII expression is linked to suppression of Sonic hedgehog ligand expression in triple negative breast cancer cells
G. Guerrini, J. Durivault, I. Filippi, M. Criscuoli, S. Monaci, J. Pouyssegur, A. Naldini, F. Carraro, S.K. Parks
Biochemical and Biophysical Research Communications.2019; 516(2): 408. CrossRef - Sonic hedgehog and Wnt/β-catenin pathways mediate curcumin inhibition of breast cancer stem cells
Xiaoting Li, Xiaoqian Wang, Chunfeng Xie, Jianyun Zhu, Yu Meng, Yue Chen, Yuan Li, Ye Jiang, Xue Yang, Shijia Wang, Jiaqi Chen, Qi Zhang, Shanshan Geng, Jieshu Wu, Caiyun Zhong, Yu Zhao
Anti-Cancer Drugs.2018; 29(3): 208. CrossRef - Glioma-Associated Oncogene Homolog Inhibitors Have the Potential of Suppressing Cancer Stem Cells of Breast Cancer
Kuo-Shyang Jeng, Chi-Juei Jeng, I-Shyan Sheen, Szu-Hua Wu, Ssu-Jung Lu, Chih-Hsuan Wang, Chiung-Fang Chang
International Journal of Molecular Sciences.2018; 19(5): 1375. CrossRef - Cancer Stem Cell Metabolism and Potential Therapeutic Targets
Vusala Snyder, Tamika C. Reed-Newman, Levi Arnold, Sufi Mary Thomas, Shrikant Anant
Frontiers in Oncology.2018;[Epub] CrossRef - Targeting the Multidrug Transporter Ptch1 Potentiates Chemotherapy Efficiency
Anida Hasanovic, Isabelle Mus-Veteau
Cells.2018; 7(8): 107. CrossRef - Combined inhibition of GLI and FLT3 signaling leads to effective anti-leukemic effects in human acute myeloid leukemia
Emily-Marie Latuske, Hauke Stamm, Marianne Klokow, Gabi Vohwinkel, Jana Muschhammer, Carsten Bokemeyer, Manfred Jücker, Maxim Kebenko, Walter Fiedler, Jasmin Wellbrock
Oncotarget.2017; 8(17): 29187. CrossRef - Prognostic role of Gli1 expression in breast cancer: a meta-analysis
Bilan Wang, Ting Yu, Yuzhu Hu, Mengmeng Xiang, Haoning Peng, Yunzhu Lin, Lu Han, Lingli Zhang
Oncotarget.2017; 8(46): 81088. CrossRef - Inhibition of Ciliogenesis Promotes Hedgehog Signaling, Tumorigenesis, and Metastasis in Breast Cancer
Nadia B. Hassounah, Martha Nunez, Colleen Fordyce, Denise Roe, Ray Nagle, Thomas Bunch, Kimberly M. McDermott
Molecular Cancer Research.2017; 15(10): 1421. CrossRef - The sonic hedgehog signaling pathway contributes to the development of salivary gland neoplasms regardless of perineural infiltration
Manuela Torres Andion Vidal, Sílvia Vanessa Lourenço, Fernando Augusto Soares, Clarissa Araújo Gurgel, Eduardo J. B. Studart, Ludmila de Faro Valverde, Iguaracyra Barreto de Oliveira Araújo, Eduardo Antônio Gonçalves Ramos, Flávia Caló de Aquino Xavier, J
Tumor Biology.2016; 37(7): 9587. CrossRef - Cancer stem cells and HER2 positive breast cancer: The story so far
Deep Shah, Clodia Osipo
Genes & Diseases.2016; 3(2): 114. CrossRef - Tamoxifen Resistance: Emerging Molecular Targets
Milena Rondón-Lagos, Victoria Villegas, Nelson Rangel, Magda Sánchez, Peter Zaphiropoulos
International Journal of Molecular Sciences.2016; 17(8): 1357. CrossRef - The Hedgehog signaling pathway is associated with poor prognosis in breast cancer patients with the CD44+/CD24− phenotype
Haishan Zhao, Hongtao Tang, Qinghuan Xiao, Miao He, Lin Zhao, Yingzi Fu, Huizhe Wu, Zhaojin Yu, Qian Jiang, Yuanyuan Yan, Feng Jin, Minjie Wei
Molecular Medicine Reports.2016; 14(6): 5261. CrossRef - Significance of the hedgehog pathway-associated proteins Gli-1 and Gli-2 and the epithelial-mesenchymal transition-associated proteins Twist and E-cadherin in hepatocellular carcinoma
Hyung Wook Chun, Ran Hong
Oncology Letters.2016; 12(3): 1753. CrossRef - Taxane‐induced hedgehog signaling is linked to expansion of breast cancer stem‐like populations after chemotherapy
Jennifer Sims‐Mourtada, Lynn M. Opdenaker, Joshua Davis, Kimberly M. Arnold, Daniel Flynn
Molecular Carcinogenesis.2015; 54(11): 1480. CrossRef - Loss of Tumor Suppressor ARID1A Protein Expression Correlates with Poor Prognosis in Patients with Primary Breast Cancer
Hyun Deuk Cho, Jong Eun Lee, Hae Yoen Jung, Mee-Hye Oh, Ji-Hye Lee, Si-Hyong Jang, Kyung-Ju Kim, Sun Wook Han, Sung Yong Kim, Han Jo Kim, Sang Byung Bae, Hyun Ju Lee
Journal of Breast Cancer.2015; 18(4): 339. CrossRef - Prognostic impact of the expression of Hedgehog proteins in cervical carcinoma FIGO stages I–IV treated with radiotherapy or chemoradiotherapy
Louise Bohr Mordhorst, Cecilia Ahlin, Bengt Sorbe
Gynecologic Oncology.2014; 135(2): 305. CrossRef - Sonic hedgehog signaling may promote invasion and metastasis of oral squamous cell carcinoma by activating MMP-9 and E-cadherin expression
Hai-Xia Fan, Shan Wang, Hong Zhao, Nian Liu, Dong Chen, Miao Sun, Jin-Hua Zheng
Medical Oncology.2014;[Epub] CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure


Fig. 1
Fig. 2
| Clinicopathologic factors | No. of cases (%) | No. of positive cases (%) |
|||||
|---|---|---|---|---|---|---|---|
| Shh | Ptch | Smo | Gli-1 | Gli-2 | Gli-3 | ||
| Age (yr) | |||||||
| < 50 | 179 (53.6) | 87 (48.6) | 100 (55.9) | 47 (26.3) | 77 (43.0) | 141 (78.8) | 99 (55.3) |
| > 50 | 155 (46.4) | 77 (49.7) | 83 (53.5) | 28 (18.1) | 64 (41.3) | 111 (71.6) | 92 (59.4) |
| p-value | 0.845 | 0.671 | 0.074 | 0.750 | 0.130 | 0.456 | |
| Size (cm) | |||||||
| < 2 | 129 (38.6) | 60 (46.5) | 72 (55.8) | 30 (23.3) | 57 (44.2) | 95 (73.6) | 85 (65.9) |
| 2-5 | 172 (51.5) | 84 (48.8) | 91 (52.9) | 38 (22.1) | 71 (41.3) | 129 (75.0) | 93 (54.1) |
| > 5 | 33 (9.9) | 20 (60.6) | 20 (60.6) | 7 (21.2) | 13 (39.4) | 28 (84.8) | 13 (39.4) |
| p-value | 0.350 | 0.687 | 0.956 | 0.829 | 0.403 | 0.011 |
|
| Node metastasis | |||||||
| 0 | 179 (53.6) | 84 (46.9) | 96 (53.6) | 40 (22.3) | 80 (44.7) | 136 (76.0) | 111 (62.0) |
| 1-3 | 98 (29.3) | 40 (40.8) | 47 (48.0) | 18 (18.4) | 36 (36.7) | 74 (75.5) | 36 (36.7) |
| > 4 | 57 (17.1) | 40 (70.2) | 40 (70.2) | 17 (29.8) | 25 (43.9) | 42 (73.7) | 44 (77.2) |
| p-value | 0.001 |
0.025 |
0.257 | 0.423 | 0.940 | 0.000 |
|
| Stage | |||||||
| I | 78 (23.4) | 32 (41.0) | 45 (57.7) | 18 (23.1) | 36 (46.2) | 57 (73.1) | 54 (69.2) |
| II | 169 (50.6) | 78 (46.2) | 83 (49.1) | 36 (21.3) | 67 (39.6) | 126 (74.6) | 89 (52.7) |
| III | 81 (24.3) | 48 (59.3) | 49 (60.5) | 19 (23.5) | 33 (40.7) | 63 (77.8) | 45 (55.6) |
| IV | 6 (1.8) | 6 (100.0) | 6 (100.0) | 2 (33.3) | 5 (83.3) | 6 (100.0) | 3 (50.0) |
| p-value | 0.007 |
0.037 |
0.896 | 0.159 | 0.475 | 0.100 | |
| Nuclear grade | |||||||
| 1 | 39 (11.7) | 20 (51.3) | 23 (59.0) | 4 (10.3) | 15 (38.5) | 34 (87.2) | 18 (46.2) |
| 2 | 168 (50.3) | 79 (47.0) | 90 (53.6) | 31 (18.5) | 68 (40.5) | 128 (76.2) | 89 (53.0) |
| 3 | 127 (38.0) | 65 (51.2) | 70 (55.1) | 40 (31.5) | 58 (45.7) | 90 (70.9) | 84 (66.1) |
| p-value | 0.747 | 0.826 | 0.004 |
0.590 | 0.112 | 0.026 |
|
| Histologic grade | |||||||
| 1 | 75 (22.5) | 34 (45.3) | 40 (53.3) | 9 (12.0) | 35 (46.7) | 61 (81.3) | 42 (56.0) |
| 2 | 136 (40.7) | 64 (47.1) | 73 (53.7) | 28 (20.6) | 61 (44.9) | 105 (77.2) | 81 (59.6) |
| 3 | 123 (36.8) | 66 (53.7) | 70 (56.9) | 38 (30.9) | 45 (36.6) | 86 (69.9) | 68 (55.3) |
| p-value | 0.433 | 0.837 | 0.007 |
0.273 | 0.160 | 0.764 | |
| ER | |||||||
| – | 154 (46.1) | 79 (51.3) | 67 (43.5) | 48 (31.2) | 53 (34.4) | 102 (66.2) | 75 (48.7) |
| + | 180 (53.9) | 85 (47.2) | 116 (64.4) | 27 (15.0) | 88 (48.9) | 150 (83.3) | 116 (64.4) |
| p-value | 0.458 | 0.000 |
0.000 |
0.008 |
0.000 |
0.004 |
|
| PR | |||||||
| – | 184 (55.1) | 95 (51.6) | 97 (52.7) | 54 (29.3) | 68 (37.0) | 139 (75.5) | 92 (50.0) |
| + | 150 (44.9) | 69 (46.0) | 86 (57.3) | 21 (14.0) | 73 (48.7) | 113 (75.3) | 99 (66.0) |
| p-value | 0.306 | 0.399 | 0.001 |
0.031 |
0.965 | 0.003 |
|
| HER2 | |||||||
| – | 251 (75.1) | 108 (43.0) | 144 (57.4) | 52 (20.7) | 107 (42.6) | 193 (76.9) | 144 (57.4) |
| + | 83 (24.9) | 56 (67.5) | 39 (47.0) | 23 (27.7) | 34 (41.0) | 59 (71.1) | 47 (56.6) |
| p-value | 0.000 |
0.099 | 0.186 | 0.790 | 0.287 | 0.905 | |
| Total (n = 290) |
LumA (n = 67) | LumB HER2 neg (n = 43) | LumB HER2 pos (n = 25) | HER2 (n = 58) | Triple negative (n = 97) | p-value | |
|---|---|---|---|---|---|---|---|
| Shh | |||||||
| – | 147 (50.7) | 33 (49.3) | 27 (62.8) | 12 (48.0) | 15 (25.9) | 60 (61.9) | 0.000 |
| + | 143 (49.3) | 34 (50.7) | 16 (37.2) | 13 (52.0) | 43 (74.1) | 37 (38.1) | |
| Ptch | |||||||
| – | 132 (45.5) | 21 (31.3) | 10 (23.3) | 13 (52.0) | 31 (53.4) | 57 (58.8) | 0.000 |
| + | 158 (54.5) | 46 (68.7) | 33 (76.7) | 12 (48.0) | 27 (46.6) | 40 (41.2) | |
| Smo | |||||||
| – | 225 (77.6) | 58 (86.6) | 40 (93.0) | 20 (80.0) | 40 (69.0) | 67 (69.1) | 0.004 |
| + | 65 (22.4) | 9 (13.4) | 3 (7.0) | 5 (20.0) | 18 (31.0) | 30 (30.9) | |
| Gli-1 | |||||||
| – | 171 (59.0) | 35 (52.2) | 25 (58.1) | 9 (36.0) | 40 (69.0) | 62 (63.9) | 0.039 |
| + | 119 (41.0) | 32 (47.8) | 18 (41.9) | 16 (64.0) | 18 (31.0) | 35 (36.1) | |
| Gli-2 | |||||||
| – | 75 (25.9) | 10 (14.9) | 8 (18.6) | 5 (20.0) | 20 (34.5) | 32 (33.0) | 0.031 |
| + | 215 (74.1) | 57 (85.1) | 35 (81.4) | 20 (80.0) | 38 (65.5) | 65 (67.0) | |
| Gli-3 | |||||||
| – | 122 (42.1) | 17 (25.4) | 20 (46.5) | 6 (24.0) | 30 (51.7) | 49 (50.5) | 0.003 |
| + | 168 (57.9) | 50 (74.6) | 23 (53.5) | 19 (76.0) | 28 (48.3) | 48 (49.5) |
| Breast carcinoma (n = 322) |
||||
|---|---|---|---|---|
| Total | No. of deceased | p-value | ||
| Age (yr) | < 50 | 170 | 17 | 0.233 |
| ≥ 50 | 152 | 19 | ||
| Stage | I | 73 | 2 | 0.000 |
| II | 163 | 14 | ||
| III | 80 | 16 | ||
| IV | 6 | 4 | ||
| Phenotype | LumA | 67 | 1 | 0.130 |
| LumB HER2 neg | 43 | 8 | ||
| LumB HER2 pos | 25 | 1 | ||
| HER2 | 58 | 5 | ||
| Triple negative | 97 | 9 | ||
| Shh | – | 167 | 17 | 0.447 |
| + | 155 | 19 | ||
| Ptch | – | 147 | 15 | 0.638 |
| + | 175 | 21 | ||
| Smo | – | 252 | 27 | 0.695 |
| + | 70 | 9 | ||
| Gli-1 | – | 186 | 25 | 0.354 |
| + | 136 | 11 | ||
| Gli-2 | – | 79 | 2 | 0.012 |
| + | 243 | 34 | ||
| Gli-3 | – | 139 | 21 | 0.469 |
| + | 183 | 15 | ||
Shh, sonic hedgehog; Ptch, patched; Smo, smoothened; Gli, glioma-associated oncogenes; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2. Statistically significant (p<0.05).
Values are presented as number (%). LumA, luminal A; LumB HER2 neg, luminal B HER2 negative; LumB HER pos, luminal B HER2 positive; Shh, sonic hedgehog; Ptch, patched; Smo, smoothened; Gli, glioma-associated oncogenes. Statistically significant (p<0.05). Classification is only available in 290 cases.
LumA, luminal A; LumB HER2 neg, luminal B HER2 negative; LumB HER pos, luminal B HER2 positive; Shh, sonic hedgehog; Ptch, patched; Smo, smoothened; Gli, glioma-associated oncogenes. Statistically significant (p<0.05).

 E-submission
E-submission