Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 47(5); 2013 > Article
-
Original Article
Diagnostic Utility of a Clonality Test for Lymphoproliferative Diseases in Koreans Using the BIOMED-2 PCR Assay - Young Kim1, Yoo Duk Choi1, Chan Choi1, Jong-Hee Nam1,2
-
Korean Journal of Pathology 2013;47(5):458-465.
DOI: https://doi.org/10.4132/KoreanJPathol.2013.47.5.458
Published online: October 25, 2013
1Department of Pathology, Chonnam National University Medical School, Gwangju, Korea.
2Brain Korea 21 Project, Center for Biomedical Human Resources at Chonnam National University, Gwangju, Korea.
- Corresponding Author: Jong-Hee Nam, M.D. Department of Pathology, Chonnam National University Hospital, Chonnam National University Medical School, 160 Baekseo-ro, Dong-gu, Gwangju 501-746, Korea. Tel: +82-62-220-4321, Fax: +82-62-227-3429, jhnam@chonnam.ac.kr
© 2013 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Background
- A clonality test for immunoglobulin (IG) and T cell receptor (TCR) is a useful adjunctive method for the diagnosis of lymphoproliferative diseases (LPDs). Recently, the BIOMED-2 multiplex polymerase chain reaction (PCR) assay has been established as a standard method for assessing the clonality of LPDs. We tested clonality in LPDs in Koreans using the BIOMED-2 multiplex PCR and compared the results with those obtained in European, Taiwanese, and Thai participants. We also evaluated the usefulness of the test as an ancillary method for diagnosing LPDs.
-
Methods
- Two hundred and nineteen specimens embedded in paraffin, including 78 B cell lymphomas, 80 T cell lymphomas and 61 cases of reactive lymphadenitis, were used for the clonality test.
-
Results
- Mature B cell malignancies showed 95.7% clonality for IG, 2.9% co-existing clonality, and 4.3% polyclonality. Mature T cell malignancies exhibited 83.8% clonality for TCR, 8.1% co-existing clonality, and 16.2% polyclonality. Reactive lymphadenitis showed 93.4% polyclonality for IG and TCR. The majority of our results were similar to those obtained in Europeans. However, the clonality for IGK of B cell malignancies and TCRG of T cell malignancies was lower in Koreans than Europeans.
-
Conclusions
- The BIOMED-2 multiplex PCR assay was a useful adjunctive method for diagnosing LPDs.
- Specimens and patients
- For this study, we selected 250 cases diagnosed with B cell lymphomas, T cell lymphomas and reactive lymphadenitis from 2004-2012 at Chonnam National University Hospital (Gwangju, Korea) and Chonnam National University Hwasun Hospital (Hwasun, Korea). The purity and concentrations of the DNA in all cases were measured. After verifying the DNA quality, 219 patients were selected and were included in clonality analyses of IG and TCR gene rearrangements, as shown in Table 1.
- The B cell lymphomas (78 cases) were comprised of 49 diffuse large B cell lymphomas (DLBCLs), 8 mucosa-associated lymphoid tissue lymphomas (MALTLs), 3 nodal marginal zone B cell lymphomas (nodal MZLs), 6 follicular lymphomas (FLs), 4 mantle cell lymphomas (MCLs), and 8 B lymphoblastic leukemias/lymphomas (BLL/L). The T cell lymphomas (80 cases) included of 41 peripheral T cell lymphomas, unspecified (PTCLs-U), 7 anaplastic large cell lymphomas (ALCLs), 12 nasal type NK/T cell lymphomas (N-NK/Ts), 12 angioimmunoblastic T cell lymphomas (AITLs), 2 adult T cell leukemia/lymphomas (ATL/Ls), and 6 T lymphoblastic leukemias/lymphomas (TLL/L). Finally, 61 patients with reactive lymphoproliferative disease included 37 incidentally-detected palpable lymphadenopathies (LAPs), 9 infection-related LAPs, 5 radiation-related LAPs, 4 autoimmune disease-related LAPs, 3 fevers of unknown origin LAPs, 2 radioactive iodine therapy-associated LAPs following surgery to treat thyroid papillary carcinomas, and 1 chemotherapy-related LAP. Two pathologists reviewed all of the hematoxylin and eosin and immunohistochemical-stained slides.
- DNA preparation
- DNA was extracted from 250 formalin-fixed, paraffin-embedded specimens. Sections (5 µm thickness) were cut from each paraffin block, and the areas of lymphoma were scraped from the unstained formalin-fixed, paraffin-embedded sections. The DNA was extracted using a QIAamp DNA Mini Kit according to the manufacturer's protocol (Qiagen, Hilden, Germany). The concentrations and purity of all 250 extracted DNA samples were measured. Polymerase chain reaction (PCR) products were obtained using a gradual dilution of DNA analyzed on MetaPhor agarose gel (Cambrex Bio Science, Rockland, ME, USA) in five cases. Bands of PCR products with DNA over 30 ng/µL were detected using the MetaPhor agarose gel. Therefore, 219 out of the 250 samples with DNA over 30 ng/µL were analyzed with the full set of BIOMED-2 PCR assay (InVivoScribe Technologies, San Diego, CA, USA) for IG and TCR gene rearrangement. Thirty-one specimens with DNA concentrations of less than 30 ng/µL were excluded from this study.
- Polymerase chain reaction analysis
- DNA volumes of 50-200 ng were used for the PCR reactions. PCR amplifications were performed using a BIOMED-2 multiplex PCR master mix (three tubes of complete IGH [IGHA: VHFR1-JH; IGHB:VHFR2-JH; IGHC:VHFR3-JH], two tubes of incomplete IGH [IGHD:DH1--6-JH; IGHE:DH7-JH], two tubes of IGK [IGKA:Vκ-Jκ; IGKB:Vκ-Kde and JκCκ intron-Kde], two tubes of complete TCRB [TCRBA:Vβ-Jβ1/2.2/2.6/2.7; TCRBB:Vβ-Jβ2.1/2.3/2.4/2.5], one tube of incomplete TCRB [TCRBC:Dβ-Jβ], and two tubes of TCRG [TCRGA:VγI/γ10-Jγ; TCRGB:Vγ9/γ11-Jγ]). The BIOMED-2 PCR protocols19 were performed using a thermal cycler according to the manufacturer's recommendations. Each PCR was carried out in a 10-µL volume (9 µL of master mix with 1 unit of AmpliTaq Gold DNA polymerase [Applied Biosystems, Foster City, CA, USA] and 1 µL of template DNA).
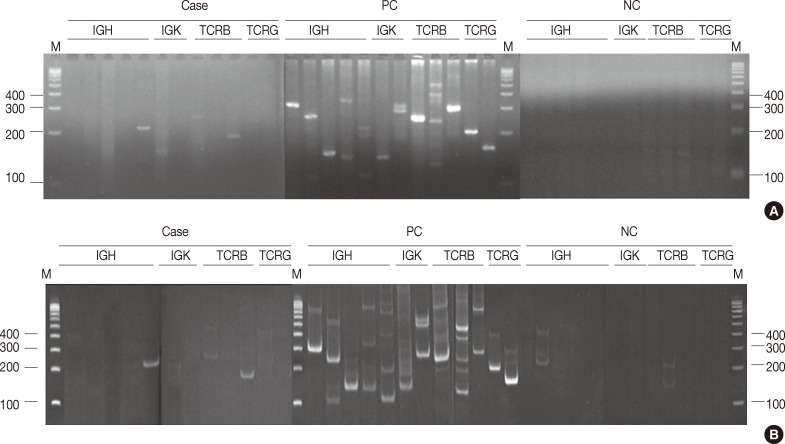
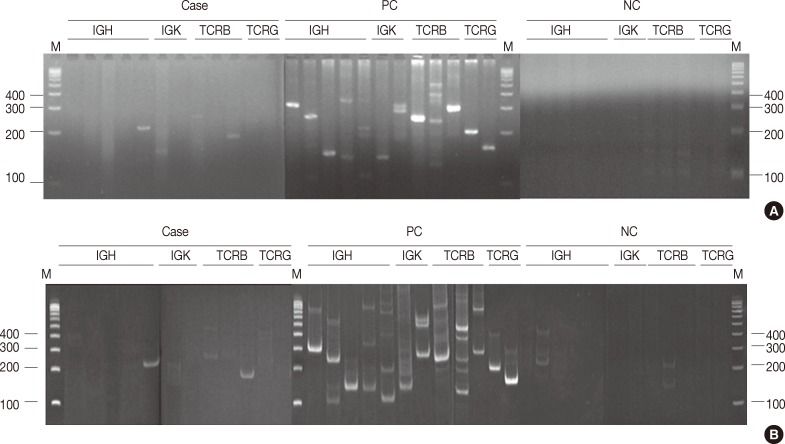
- PCR products were denatured at 95℃ for 10 minutes and rapidly transferred to 4℃ for 60 minutes to promote duplex formation. Next, 480 reactions (40 cases) were analyzed on both 8% non-denaturing polyacrylamide (acrylamide:bisacrylamide=29:1) gel in a 1× Tris-boric acid-EDTA buffer, and on 4% MetaPhor agarose gel in a 1× Tris-boric acid-EDTA buffer at 100 V for 50 minutes simultaneously. The other reactions were analyzed on MetaPhor agarose gel only.
- Interpretation of clonality
- Gel images were examined independently by two pathologists experienced in clonality analysis. When the PCR products appeared indistinct and one or more ill-defined bands of varying size ranges were present, the PCR products were interpreted as polyclonal. The rearrangements were interpreted as clonal when one or more discrete bands appeared within the expected size range, and they were often accompanied by one or more bands in IG or TCR.20 Co-existing clonality was defined when one or more discrete bands within the expected size range appeared in IG and TCR. Co-existing clonality was regarded as a subtype of clonality in the current study. In cases with weak PCR bands or equivocal results, the amplification was repeated and the amplicons were analyzed on both 8% non-denaturing polyacrylamide gel and 4% MetaPhor agarose gel.
MATERIALS AND METHODS
- Among 70 cases of mature B cell lymphomas, 67 (95.7%) showed clonality for IG, 2 (2.9%) exhibited co-existing clonality for IG and TCR, and 3 (4.3%) showed polyclonality. Four out of eight (50%) BLL/L cases revealed clonality for IG, five cases (62.5%) had clonality for TCR, two cases (25%) exhibited co-existing clonality, and one (12.5%) had polyclonality. Sixty-two out of 74 cases (83.8%) diagnosed with mature T cell lymphoma revealed clonality for TCR, 12 cases (16.2%) showed polyclonality, and 6 (8.1%) cases revealed co-existing clonality. Four out of six TLL/L cases showed clonality for TCR, and two cases showed polyclonality. Among 61 cases of reactive lymphadenitis, 57 cases (93.4%) were polyclonal for IG and TCR; however, 4 cases (6.6%) had clonality for IGK or TCRG. Two cases (3.3%) were clonal for each IGKA and TCRGB (Table 2).
- Mature B cell lymphoma revealed clonality for IGH in 60 out of total 70 cases. Among 70 mature B cell lymphoma cases, 56 cases (64.0%) had clonality for the VDJ region of IGH and 35 (50.0%) revealed clonality for the DH-JH region of IGH. Thirty-three cases (47.1%) showed clonality for IGK. Sixty-seven out of 70 (95.7%) cases had clonality for IGH or IGK. The DLBCLs revealed clonality for IGH in 41 out of 49 (83.7%) cases, including 38 (77.6%) that showed clonality for the VDJ region and 29 (59.2%) for the DH-JH region. Finally, 18 (36.7%) cases had clonality for IGK. Two cases of DLBCL showed combined TCR gene rearrangement, and three cases revealed polyclonality. Other mature B cell lymphomas showed 100% clonality for IG. Of the cases with MALTLs, eight (100%) had clonality for IGH, and four (50.0%) had clonality for IGK. Of those cases with nodal MZLs, three (100%) had clonality for IGH and two (66.7%) showed clonality for IGK. Three of the four cases with MCLs (75.0%) had clonality for IGH and all 4 (100%) had clonality for IGK. All FL cases showed clonality for IGH or IGK, but no clonality for the DH-JH region of IGH was found. BLL/L demonstrated quite different results compared to other B cell lymphomas. BLL/L revealed four cases (50%) with clonality for IG, including three (37.5%) that showed clonality for IGH and one (12.5%) with clonality for IGK. Five out of eight BLL/L showed TCR gene rearrangements including two TCRB clonalities and/or five TCRG clonalities. Co-existing clonal cases were two and polyclonal case was one in BLL/L (Table 3).
- Among 74 mature T cell lymphomas, 62 cases (83.8%) had TCR clonality. There were 38 clonal cases (51.4%) each for TCRB and TCRG (Table 4). Clonal bands were observed in 23 (31.1%) and 26 cases (35.1%) for the Vβ-Jβ and Dβ-Jβ regions of TCRB, respectively. Thirty-seven of 41 (90.2%) PTCL-U cases had clonality for TCR, including 19 cases (46.3%) with clonality for TCRB and 21 cases (51.2%) with clonality for TCRG. N-NK/Ts had clonality for TCR in 8 cases (66.7%). AITL had clonality in 10 (83.3%), and ATL/Ls had clonality for TCR in 2 (100%). N-NK/Ts had clonality for TCRB in seven cases (58.3%) and for TCRG in four cases (33.3%). TCRB and TCRG gene rearrangements of AITL were revealed in six cases (50%) and eight cases (66.7%), respectively. ALCLs had clonality for total TCR in five cases (71.4%) including four cases (57.1%) of clonality for TCRB and three cases (42.9%) for TCRG. All TLL/L cases had no clonality for IGH or IGK, and four out of six (66.7%) TLL/L showed TCR clonality, including two TCRB and two TCRG clonal cases (Table 4).
- In all, 480 reactions spanning 40 cases were analyzed on non-denaturing polyacrylamide gels and MetaPhor agarose gels (Cambrex Bio Science) simultaneously. All 480 reactions revealed the same results (Fig. 1A, B).
RESULTS
- The clonality detection rate of LPDs of Koreans is lower than that of Europeans but higher than that of the Taiwanese. All mature B cell lymphomas in this study from a Korean population showed 67 out of 70 cases (95.7%) of clonality for the IG gene compared to 367 out of 369 (99.0%) in a European study.21 Twenty-two cases (95.7%) in B cell lymphomas of southern Taiwan had clonality for IG, which was the same finding as our data.17 Sung et al.'s study22 in Koreans using BIOMED-2 multiplex PCR reported 87.5% (14/16) clonality for IG. Our cases showed clonality for IGH in 60 out of 70 cases (85.7%), whereas for IGK, clonality was shown in 33 out of 70 cases (47.1%). IGK clonality was significantly lower compared to that in a European patient sample, which revealed 323 out of 369 cases (88.0%) (p<.05) of clonality, and it was also lower compared to that in a Taiwanese patient sample, which found 16 of 23 cases (69.6%) of clonality.17,21 However, in a Korean patient sample, Sung et al.22 reported that 7 of 16 cases (43.8%) showed clonality for IGK, which was similar to our findings. Of the DLBCL cases, 93.8% showed clonality for IG, which was similar to a European study that revealed 98.0% clonality.15 The other types of B cell lymphomas showed 100% clonality, similar to what was observed in a European study.14,15 The FL cases showed 100% (6/6) clonality for IG including IGH and IGK; however, no clonality was observed for DH-JH rearrangements of IGH in this study, as compared to a 19.0% rearrangement rate in a European study.15
- BLL/L showed quite different clonality results from other types of B cell lymphomas. BLL/L had clonality for IG in four of eight cases (50%), which was much lower than other types of B cell lymphomas, and clonality for TCR in five of eight cases (62.5%), which was much higher than other types of B cell lymphomas. In addition, co-existing clonality was presented in two cases (25%) in BLL/L, which was higher than other B cell lymphomas except DLBCL. This finding might be because BLL/L are arrested at various stages of pre-B cell development and cross lineage IG/TCR gene rearrangement occurs particularly in immature lymphoid malignancy. Therefore, in the case of LPD of unknown origin, the IG/TCR gene rearrangement result should not be used directly for determination of B or T cell lineage.15
- There were three polyclonal cases out of 70 (4.3%) B cell lymphomas, all of which were DLBCLs. The ratio of polyclonal B cell lymphomas was higher among the Korean sample than the European sample, which revealed polyclonality in two DLBCLs out of 369 B cell lymphomas. There was one polyclonal case out of 23 B cell lymphomas in Taiwan, which was also DLBCL. Polyclonality may occur in lymphoid malignancies when the number of clonal B cells is too low to be detected or the cases have a significant somatic hypermutation without gene rearrangement.21
- Moreover, there were two co-existing clonal cases in DLBCLs and BLL/L, respectively. One case out of two co-existing DLBCLs showed bands in IGKA and TCRBc, while the other case revealed discrete bands in IGHC, IGHD, IGKA, and TCRGA. One case of BLL/L was positive for IGKB and TCRGB, and the other case showed bands in all tubes of IGH, TCRBA and TCRBB, and TCRGB. The study of Europeans reported that co-existing clonality was 27% in mature B cell lymphomas. Additionally, the incidence of cross lineage IG/TCR gene rearrangement has been reported to be up to 90% in BLL/L.21 Although the causes are currently not well understood, the rearrangements might arise from somatic hypermutations or point mutations.21
- The clonality of T cell lymphomas in this study was lower than that of Europeans but higher than that of the Taiwanese. The clonality of all T cell lymphomas except N-NK/T and TLL/L in this study was 87.1% (54/62), as compared to 94.0% (177/188) in a European study and 76.0% (19/25) in a Taiwanese study.12,17 Among T cell lymphomas, including N-NK/T and TLL/L, TCR gene rearrangement was revealed in 66 out of 80 cases (82.5%) in this study. There was clonality for TCRB in 38 out of 74 cases (51.4%) mature T cell lymphomas in Koreans compared with 171 out of 188 cases (91.0%) in a European study and 11 out of 28 cases (39.3%) in a Taiwanese study. Additionally, clonality for TCRG was found in 38 cases (51.4%) in this study, which was significantly lower than the data reported in the European-based study (168/188, 89.0%; p<.05) and similar to that of the Taiwanese study (17/29, 58.6%).12,14,15,17 In PTCL-U cases, 21 out of 41 cases (51.2%) showed clonality for TCRG compared with 177 out of 188 (94.0%) in a European study and 10 out of 14 (71.4%) in a Taiwanese study. One of the reasons for this difference might be ethnic differences. Europeans have minor allele frequencies of 0.33, whereas African Americans have frequencies of 0.50, and Asians 0.42 for the allele of a single nucleotide polymorphism (rs 2392542) for TCRG.23 N-NK/T showed clonality for TCR in eight out of 12 (66.7%), which was higher than those of a Thai group (20/67, 30%) and a Taiwan study (0/8, 0%).17,18 Of seven ALCL cases, five (71.4%) had clonality for TCR, which was similar to data from a European study that reported clonality in 34 out of 43 (79.0%).12 Of 12 AITL cases, 10 (83.3%) had clonality for TCR compared with 35 out of 37 (95.0%) in the European data.12
- Twelve polyclonal cases were observed in 74 T cell lymphomas. They were four PTCLs-U, four N-NK/Ts, two AITLs, and two anaplastic lymphoma kinase (ALK)-positive ALCLs. The European study revealed 11 polyclonal cases in 188 (5.9%) T cell lymphomas, which consisted of eight ALK-positive ALCLs, one ALK-negative ALCL, and two AITLs. There were no polyclonal cases for TCR among PTCLs-U in a European study. European data reported that 24.0% of ALK-positive ALCLs did not demonstrate TCR clonality.12
- Four PTCL-U with co-existing clonality showed various IG and TCR gene rearrangements, which were IGHC and IGHD, IGKB, TCRBA and TCRBB, and TCRGB. Two AITLs revealed co-existing clonality for IGHB, IGHC, IGHD, IGKB, and TCRBA. In this study, six cases (8.1%) with co-existing clonality were identified in 74 mature T cell lymphomas. The rate of 0-9% for co-existing clonality has been reported in T cell lymphomas of Europeans.12 Four out of six (66.7%) TLL/L showed clonality for TCR, which is similar to that of other T cell lymphomas. In contrast to BLL/L, all of the TLL/L cases showed no co-existing clonality.
- Most of the 61 cases diagnosed with reactive LPD had polyclonality (93.4%). However, four out of 61 cases (6.6%) with reactive lymphadenitis had clonality as well. A case with clonality for IGK involved a 53-year-old female with inguinal LAP who had a history of radiation therapy due to uterine cervical cancer. The clonality may have resulted from the radiation therapy, as previously reported.24 The second case with clonality for IGK was a 28-year-old male who was infected with human immunodeficiency virus (HIV). It has been reported that IG rearrangements are associated with HIV-related LAP syndrome.20 The third case with clonality for TCRG was a 53-year-old male who had received radioactive iodine therapy after a total thyroidectomy due to papillary thyroid carcinoma. Radioactive iodine therapy is reported to cause clonality of the lymph nodes.25 The fourth case was a 37-year-old female with a history of virus-induced cervical LAP. The clonality for TCRG may have been induced by a viral infection in this case.20 There was no occurrence of lymphoma in these cases during the three-year follow-up period.
- Cho et al.8 reported on IG and TCR gene rearrangements in LPDs based on a conventional nested PCR-based clonality assay using a FRIIIA-LJH/VLJH consensus primer for IGH and a Vγ-Jγ consensus primer for TCRG in a Korean population. The clonality for IGH in B cell lymphomas was 97.1% (34/35), which was higher than the 85.7% clonality in the current study. However, the specificity of IGH gene rearrangements was 97%, which was lower than the 100% specificity in BIOMED-2 found in this study. In addition, the clonality for TCRG in T cell lymphoma was 79.3%,8 which was higher than the 51.2% (21/41) reported herein. The specificity of TCRG gene rearrangements using the previous method was 99.4%, which was higher than the 96.7% found by the current study. The higher clonality ratio for IG and TCR might result from the semi-nested PCR technique, which runs the PCR twice.
- Electrophoresis of 480 PCR products was performed using 4% MetaPhor agarose gels and 8% non-denaturing polyacrylamide gels, which revealed the same results on both gels. Although the manufacturer of the BIOMED-2 multiplex PCR kit recommends electrophoresis of the PCR products using a non-denaturing polyacrylamide gel or gene scan, the MetaPhor agarose gel is as sensitive as a non-denaturing polyacrylamide gel. MetaPhor agarose gel has several advantages over non-denaturing polyacrylamide gel, such as ease of handling, safe and quick preparation. Therefore, all of the PCR products in this study were analyzed using 4% MetaPhor agarose gels. In cases with weak PCR bands or equivocal results, the analysis of both 8% non-denaturing polyacrylamide gel and 4% MetaPhor agarose gel will help confirm the diagnosis.
- This study had some limitations. We performed IGH, IGK, TCRB, and TCRG tests, but the PCR amplification for IGL and TCRD was not carried out. It has been reported that the effect of IGL or TCRD testing for clonality detection is negligible, ranging from1 to 2%.12,21 Sung et al.22 also found a similar result. They observed the clonality detection rate using IGH and IGK was not changed irrespective of the IGL test. Additionally, the number of cases in this study was less than that of the European study. More cases of Koreans would be of help in comparing among races and establishing a definitive diagnosis.
- Although there were some differences in clonalities for IGK and TCRG of B and T cell lymphomas between Korean and European populations, the overall clonality ratio in B or T cell lymphomas of Koreans was similar to those of Europeans. We conclude that the BIOMED-2 multiplex PCR reaction can be a useful adjuvant test for diagnosing LPDs when the histologic and immunohistochemical evidence is insufficient.
DISCUSSION
Acknowledgments
Acknowledgments
- 1. National Cancer Informantion Center now covers statistics of cancer prevalence [Internet]. Goyang: National Cancer Information Center, c2012; cited 2013 Sep 1. Available from: http://www.cancer.go.kr/mbs/cancer/subview.jsp?id=cancer_040102000000.
- 2. Gleissner B, Maurer J, Thiel E. Detection of immunoglobulin heavy chain genes rearrangements in B-cell leukemias, lymphomas, multiple myelomas, monoclonal and polyclonal gammopathies. Leuk Lymphoma 2000; 39: 151-155. ArticlePubMed
- 3. Langerak AW, van Krieken JH, Wolvers-Tettero IL, et al. The role of molecular analysis of immunoglobulin and T cell receptor gene rearrangements in the diagnosis of lymphoproliferative disorders. J Clin Pathol 2001; 54: 565-567. ArticlePubMedPMC
- 4. Sen F, Vega F, Medeiros LJ. Molecular genetic methods in the diagnosis of hematologic neoplasms. Semin Diagn Pathol 2002; 19: 72-93. PubMed
- 5. Sioutos N, Bagg A, Michaud GY, et al. Polymerase chain reaction versus Southern blot hybridization: detection of immunoglobulin heavy-chain gene rearrangements. Diagn Mol Pathol 1995; 4: 8-13. ArticlePubMed
- 6. Achille A, Scarpa A, Montresor M, et al. Routine application of polymerase chain reaction in the diagnosis of monoclonality of B-cell lymphoid proliferations. Diagn Mol Pathol 1995; 4: 14-24. ArticlePubMed
- 7. Benhattar J, Delacretaz F, Martin P, Chaubert P, Costa J. Improved polymerase chain reaction detection of clonal T-cell lymphoid neoplasms. Diagn Mol Pathol 1995; 4: 108-112. ArticlePubMed
- 8. Cho EY, Ko YH, Kim DS, Han JJ, Ree HJ. Diagnostic utility of polymerase chain reaction-based clonality analysis for immunoglobulin heavy chain gene and T-cell receptor gamma chain gene rearrangement in lymphoid neoplasms. Korean J Pathol 2001; 35: 461-469.
- 9. Fodinger M, Buchmayer H, Schwarzinger I, et al. Multiplex PCR for rapid detection of T-cell receptor-gamma chain gene rearrangements in patients with lymphoproliferative diseases. Br J Haematol 1996; 94: 136-139. ArticlePubMedPDF
- 10. Ilyas M, Jalal H, Linton C, Rooney N. The use of the polymerase chain reaction in the diagnosis of B-cell lymphomas from formalin-fixed paraffin-embedded tissue. Histopathology 1995; 26: 333-338. ArticlePubMed
- 11. Lozano MD, Tierens A, Greiner TC, Wickert RS, Weisenburger DD, Chan WC. Clonality analysis of B-lymphoid proliferations using the polymerase chain reaction. Cancer 1996; 77: 1349-1355. ArticlePubMed
- 12. Brüggemann M, White H, Gaulard P, et al. Powerful strategy for polymerase chain reaction-based clonality assessment in T-cell malignancies Report of the BIOMED-2 Concerted Action BHM4 CT98-3936. Leukemia 2007; 21: 215-221. ArticlePubMedPDF
- 13. Droese J, Langerak AW, Groenen PJ, et al. Validation of BIOMED-2 multiplex PCR tubes for detection of TCRB gene rearrangements in T-cell malignancies. Leukemia 2004; 18: 1531-1538. ArticlePubMedPDF
- 14. Liu H, Bench AJ, Bacon CM, et al. A practical strategy for the routine use of BIOMED-2 PCR assays for detection of B- and T-cell clonality in diagnostic haematopathology. Br J Haematol 2007; 138: 31-43. ArticlePubMed
- 15. van Krieken JH, Langerak AW, Macintyre EA, et al. Improved reliability of lymphoma diagnostics via PCR-based clonality testing: report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia 2007; 21: 201-206. ArticlePubMedPDF
- 16. McClure RF, Kaur P, Pagel E, et al. Validation of immunoglobulin gene rearrangement detection by PCR using commercially available BIOMED-2 primers. Leukemia 2006; 20: 176-179. ArticlePubMedPDF
- 17. Chen YL, Su IJ, Cheng HY, et al. BIOMED-2 protocols to detect clonal immunoglobulin and T-cell receptor gene rearrangements in B- and T-cell lymphomas in southern Taiwan. Leuk Lymphoma 2010; 51: 650-655. ArticlePubMed
- 18. Pongpruttipan T, Sukpanichnant S, Assanasen T, et al. Extranodal NK/T-cell lymphoma, nasal type, includes cases of natural killer cell and alphabeta, gammadelta, and alphabeta/gammadelta T-cell origin: a comprehensive clinicopathologic and phenotypic study. Am J Surg Pathol 2012; 36: 481-499. PubMed
- 19. van Dongen JJ, Langerak AW, Brüggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-C T98-3936. Leukemia 2003; 17: 2257-2317. ArticlePubMedPDF
- 20. Langerak AW, Molina TJ, Lavender FL, et al. Polymerase chain reaction-based clonality testing in tissue samples with reactive lymphoproliferations: usefulness and pitfalls: a report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2007; 21: 222-229. ArticlePubMedPDF
- 21. Evans PA, Pott C, Groenen PJ, et al. Significantly improved PCR-based clonality testing in B-cell malignancies by use of multiple immunoglobulin gene targets. Report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia 2007; 21: 207-214. ArticlePubMedPDF
- 22. Sung JY, Kang SY, Kim SH, Kwon JE, Ko YH. Analysis of immunoglobulin gene rearrangement: comparison between BIOMED-2 multiplex PCR and conventional nested PCR. Lab Med Online 2011; 1: 195-201. Article
- 23. National Center for Biotechnology Information now covers SNP [Internet]. Bethesda: National Center for Biotechnology Information, cited 2013 Sep 1. Available from: http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.
- 24. Fairhurst RM, Valles-Ayoub Y, Neshat M, Braun J. A DNA repair abnormality specific for rearranged immunoglobulin variable genes in germinal center B cells. Mol Immunol 1996; 33: 231-244. ArticlePubMed
- 25. Vershenya S, Biko J, Drozd V, Lorenz R, Reiners C, Hempel K. Dose response for T-cell receptor (TCR) mutants in patients repeatedly treated with 131I for thyroid cancer. Mutat Res 2004; 548: 27-33. ArticlePubMed
REFERENCES
NAValues are presented as number (%).
NHL, non-Hodgkin lymphoma; RLD, reactive lymphoproliferative disease; MCL, mantle cell lymphoma; NA, not applicable; FL, follicular lymphoma; MZL, marginal zone lymphoma; MALTL, extranodal marginal zone lymphoma of mucosa associated lymphoid tissue; DLBCL, diffuse large B-cell lymphoma; BLL/L, B lymphoblastic lymphoma/leukemia; PTCL-U, peripheral T cell lymphoma, unspecified; ALCL, anaplastic large cell lymphoma; N-NK/T, nasal type NK/T cell lymphoma; AITL, angioimmunoblastic T-cell lymphoma; ATL/L, adult T cell leukemia/lymphoma; TLL/L, T lymphoblastic leukemia/lymphoma.
Values are presented as number (%).
MCL, mantle cell lymphoma; FL, follicular lymphoma; MZL, marginal zone lymphoma; MALTL, extranodal marginal zone lymphoma of mucosa associated lymphoid tissue; DLBCL, diffuse large B-cell lymphoma; BLL/L, B lymphoblastic leukemia/lymphoma; IG, immunoglobulin; TCR, T cell receptor.
TCL, T cell lymphoma; PTCL-U, peripheral T cell lymphoma, unspecified; ALCL, anaplastic large cell lymphoma; N-NK/T, nasal type NK/T cell lymphoma; AITL, angioimmunoblastic T-cell lymphoma; ATL/L, adult T cell leukemia/lymphoma; TLL/L, T lymphoblastic leukemia/lymphoma; TCR, T cell receptor; IG, immunoglobulin
Figure & Data
References
Citations

- Infectious mononucleosis complicated by transitory Epstein-Barr virus infection of T and natural killer cells
Yanlin Zhang, JianLan Xie, Yuanyuan Zheng, XiaoGe Zhou
Journal of Hematopathology.2024; 17(3): 129. CrossRef - Experiencia en el uso de protocolos Biomed-2 para el estudio de reordenamientos de TCR e inmunoglobulinas en proliferaciones linfoides en el Instituto Nacional de Cancerología, Colombia
Nicolás Villamizar-Rivera, Natalia Olaya
Biomédica.2022; 42(Sp. 1): 64. CrossRef - Enhancing diagnosis of T-cell lymphoma using non-recombined T-cell receptor sequences
Yi-Lin Chen, Chung-Liang Ho, Chen-Yan Hung, Wan-Li Chen, Chen Chang, Yi-Hsin Hou, Jian-Rong Chen, Pin-Jun Chen, Nan-Haw Chow, Wenya Huang, Ya-Ting Hsu, Tsai-Yun Chen, Tsunglin Liu
Frontiers in Oncology.2022;[Epub] CrossRef - The utility and limitations of B- and T-cell gene rearrangement studies in evaluating lymphoproliferative disorders
Hadrian Mendoza, Christopher A. Tormey, Henry M. Rinder, John G. Howe, Alexa J. Siddon
Pathology.2021; 53(2): 157. CrossRef - Combined detection of lymphocyte clonality and MALT1 translocations in bronchoalveolar lavage fluid for diagnosing pulmonary lymphomas
Takashi Kido, Hiroshi Ishimoto, Hiroshi Ishii, Kanako Hara, Mutsumi Ozasa, Hiroki Kawabata, Toshinori Kawanami, Yu Suzuki, Hiroki Yoshikawa, Atsuko Hara, Noriho Sakamoto, Nobuhiro Matsumoto, Chiharu Yoshii, Junya Fukuoka, Masaki Fujita, Masamitsu Nakazato
Scientific Reports.2021;[Epub] CrossRef - Differentiation of lymphocytic-plasmacytic enteropathy and small cell lymphoma in cats using histology-guided mass spectrometry
Sina Marsilio, Shelley J. Newman, James Scot Estep, Paula R. Giaretta, Jonathan A. Lidbury, Emma Warry, Andi Flory, Paul S. Morley, Katy Smoot, Erin H. Seeley, Matthew J. Powell, Jan S. Suchodolski, Jörg M. Steiner
Journal of Veterinary Internal Medicine.2020; 34(2): 669. CrossRef - T-Cell Receptor Rearrangements Determined Using Fragment Analysis in Patients With T-Acute Lymphoblastic Leukemia
Hyerim Kim, In-Suk Kim, Chulhun L. Chang, Sun-Young Kong, Young Tak Lim, Seom Gim Kong, Eun Hae Cho, Eun-Yup Lee, Ho-Jin Shin, Hyeon Jin Park, Hyeon-Seok Eom, Hyewon Lee
Annals of Laboratory Medicine.2019; 39(2): 125. CrossRef - Monitoring immunoglobulin heavy chain and T‑cell receptor gene rearrangement in cfDNA as minimal residual disease detection for patients with acute myeloid leukemia
Ling Zhong, Jiao Chen, Xiaobing Huang, Yanxing Li, Tao Jiang
Oncology Letters.2018;[Epub] CrossRef - Molecular pathology diagnosis of diffuse large B cell lymphoma using BIOMED-2 clonal gene rearrangements
Saeid Ghorbian
Annals of Diagnostic Pathology.2017; 29: 28. CrossRef - Improved clonality detection in B‐cell lymphoma using a semi‐nested modification of the BIOMED‐2 PCR assay for IGH rearrangement: A paraffin‐embedded tissue study
Yuma Sakamoto, Ayako Masaki, Satsuki Aoyama, Shusen Han, Kosuke Saida, Kana Fujii, Hisashi Takino, Takayuki Murase, Shinsuke Iida, Hiroshi Inagaki
Pathology International.2017; 67(9): 453. CrossRef - The prognostic significance of monoclonal immunoglobulin gene rearrangement in conjunction with histologic B‐cell aggregates in the bone marrow of patients with diffuse large B‐cell lymphoma
Yoon Ah Cho, Woo Ick Yang, Jae‐Woo Song, Yoo Hong Min, Sun Och Yoon
Cancer Medicine.2016; 5(6): 1066. CrossRef - Nasal-type NK/T-cell lymphomas are more frequently T rather than NK lineage based on T-cell receptor gene, RNA, and protein studies: lineage does not predict clinical behavior
Mineui Hong, Taehee Lee, So Young Kang, Suk-Jin Kim, Wonseog Kim, Young-Hyeh Ko
Modern Pathology.2016; 29(5): 430. CrossRef - Long-term Tumor-free Survival With Untreated Primary Diffuse Large B-cell Lymphoma of the Tonsil
Xiaojing Zhang, Yuanyuan Zheng, Jianlan Xie, Jun Zhu, Yuqin Song, Xiaojing Teng, Wei Liu, Yi Ding, Yuhua Huang, Xiaoge Zhou
American Journal of Surgical Pathology.2015; 39(11): 1493. CrossRef - Evaluation diagnostic usefulness of immunoglobulin light chains (Igκ, Igλ) and incomplete IGH D-J clonal gene rearrangements in patients with B-cell non-Hodgkin lymphomas using BIOMED-2 protocol
S. Ghorbian, I. Jahanzad, G. R. Javadi, E. Sakhinia
Clinical and Translational Oncology.2014; 16(11): 1006. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure

Fig. 1
| Characteristic | NHL | RLD |
|---|---|---|
| No. | 158 (100) | 61 (100) |
| Age (yr) | ||
| < 30 | 21 (13.3) | 12 (19.7) |
| 30-49 | 27 (17.1) | 19 (31.1) |
| 50-64 | 49 (31.0) | 18 (29.5) |
| > 65 | 61 (38.6) | 12 (19.7) |
| Mean (range) | 57.8 (7-88) | 50.8 (3-78) |
| Sex | ||
| Male | 94 | 36 |
| Female | 64 | 25 |
| Subtype | ||
| MCL | 4 | NA |
| FL | 6 | NA |
| MZL | 3 | NA |
| MALTL | 8 | NA |
| DLBCL | 49 | NA |
| BLL/L | 8 | NA |
| PTCL-U | 41 | NA |
| ALCL | 7 | NA |
| N-NK/T | 12 | NA |
| AITL | 12 | NA |
| ATL/L | 2 | NA |
| TLL/L | 6 | NA |
| BCL (n = 78) |
TCL (n = 80) |
RLD (n = 61) | |||
|---|---|---|---|---|---|
| Mature BCL (n = 70) | BLL/L (n = 8) | Mature TCL (n = 74) | TLL/L (n = 6) | ||
| IGH+IGK | 67 (95.7) | 4 (50.0) | 6 (8.1) | 0 (0) | 2 (3.3) |
| TCRB+TCRG | 2 (2.9) | 5 (62.5) | 62 (83.8) | 4 (66.7) | 2 (3.3) |
| Co-existing clonality | 2 (2.9) | 2 (25.0) | 6 (8.1) | 0 (0) | 0 (0) |
| Polyclonality | 3 (4.3) | 1 (12.5) | 12 (16.2) | 2 (33.3) | 57 (93.4) |
| Mature BCL |
Precursor BCL |
Total (n = 78) | ||||||
|---|---|---|---|---|---|---|---|---|
| MCL (n = 4) | FL (n = 6) | MZL (n = 3) | MALTL (n = 8) | DLBCL (n = 49) | Subtotal (n = 70) | BLL/L (n = 8) | ||
| IGH genes | ||||||||
| No rearrangement | 1 (25.0) | 1 (16.7) | 0 (0) | 0 (0) | 8 (16.3) | 10 (4.3) | 5 (62.5) | 15 (23.1) |
| VDJ cases | 2 (50.0) | 5 (83.3) | 3 (100) | 8 (100) | 38 (77.6) | 56 (64.0) | 3 (37.5) | 59 (75.6) |
| DH-JH cases | 1 (25.0) | 0 (0) | 1 (33.3) | 4 (50.0) | 29 (59.2) | 35 (50.0) | 1 (12.5) | 36 (46.2) |
| All IGH VDJ+DH | 3 (75.0) | 5 (83.3) | 3 (100) | 8 (100) | 41 (83.7) | 60 (85.7) | 3 (37.5) | 63 (80.8) |
| IGK genes | ||||||||
| No rearrangement | 0 (0) | 1 (16.7) | 1 (33.3) | 4 (50.0) | 31 (63.3) | 37 (52.9) | 7 (87.5) | 44 (56.4) |
| Total IGK | 4 (100) | 5 (83.3) | 2 (66.7) | 4 (50.0) | 18 (36.7) | 33 (47.1) | 1 (12.5) | 34 (43.6) |
| Combined IG genes | ||||||||
| All IGH+IGK | 4 (100) | 6 (100) | 3 (100) | 8 (100) | 46 (93.8) | 67 (95.7) | 4 (50.0) | 71 (91.0) |
| TCRB genes | ||||||||
| Vβ-Jβ genes | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (2.0) | 1 (1.4) | 1 (12.5) | 2 (2.6) |
| Dβ-Jβ genes | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (12.5) | 1 (1.3) |
| Total TCRB | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (2.0) | 1 (1.4) | 2 (25.0) | 3 (3.8) |
| TCRG genes | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (2.0) | 1 (1.4) | 5 (62.5) | 6 (7.7) |
| Combined TCR genes | ||||||||
| All TCRB+TCRG | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (4.1) | 2 (2.9) | 5 (62.5) | 7 (9.0) |
| Polyclonality | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (6.1) | 3 (6.1) | 1 (12.5) | 4 (5.1) |
| Mature TCL |
Precursor TCL |
Total (n = 80) | ||||||
|---|---|---|---|---|---|---|---|---|
| PTCL-U (n = 41) | ACLC (n = 7) | N-NK/T (n = 12) | AITL (n = 12) | ATL/L (n = 2) | Subtotal (n = 74) | TLL/L (n = 6) | ||
| TCRB genes | ||||||||
| No rearrangement | 22 (53.7) | 4 (57.1) | 5 (41.7) | 5 (41.7) | 0 (0) | 36 (48.6) | 4 (75.0) | 40 (50.0) |
| Vβ-Jβ cases | 12 (29.3) | 2 (28.6) | 2 (16.7) | 5 (41.7) | 2 (100) | 23 (31.1) | 1 (16.7) | 24 (30.0) |
| Dβ-Jβ cases | 13 (31.7) | 4 (57.1) | 5 (41.7) | 3 (25.0) | 1 (50.0) | 26 (35.1) | 1 (16.7) | 27 (33.8) |
| Total TCRB | 19 (46.3) | 4 (57.1) | 7 (58.3) | 6 (50.0) | 2 (100) | 38 (51.4) | 2 (33.3) | 40 (50.0) |
| TCRG genes | ||||||||
| No rearrangement | 19 (46.3) | 4 (57.1) | 8 (66.7) | 4 (33.3) | 0 (0) | 35 (47.3) | 4 (66.7) | 39 (48.8) |
| Total TCRG | 21 (51.2) | 3 (42.9) | 4 (33.3) | 8 (66.7) | 2 (100) | 38 (51.4) | 2 (33.3) | 40 (50.0) |
| Total TCR | 37 (90.2) | 5 (71.4) | 8 (66.7) | 10 (83.3) | 2 (100) | 62 (83.8) | 4 (66.7) | 66 (82.5) |
| IGH genes | ||||||||
| VDJ cases | 1 (2.4) | 0 (0) | 0 (0) | 2 (16.7) | 0 (0) | 3 (4.0) | 0 (0) | 3 (3.8) |
| DH-JH cases | 3 (7.3) | 0 (0) | 0 (0) | 1 (8.3) | 0 (0) | 4 (5.4) | 0 (0) | 4 (5.0) |
| All IGH VDJ+DH | 3 (7.3) | 0 (0) | 0 (0) | 2 (16.7) | 0 (0) | 5 (6.8) | 0 (0) | 5 (6.3) |
| IGK genes | 1 (2.4) | 0 (0) | 0 (0) | 1 (8.3) | 0 (0) | 2 (2.7) | 0 (0) | 2 (2.5) |
| Combined IG genes | ||||||||
| All IGH+IGK | 3 (7.3) | 0 (0) | 0 (0) | 2 (16.7) | 0 (0) | 5 (6.8) | 0 (0) | 5 (6.3) |
| Polyclonality | 4 (9.8) | 2 (28.6) | 4 (33.3) | 2 (16.7) | 0 (0) | 12 (16.2) | 2 (33.3) | 14 (17.5) |
NAValues are presented as number (%). NHL, non-Hodgkin lymphoma; RLD, reactive lymphoproliferative disease; MCL, mantle cell lymphoma; NA, not applicable; FL, follicular lymphoma; MZL, marginal zone lymphoma; MALTL, extranodal marginal zone lymphoma of mucosa associated lymphoid tissue; DLBCL, diffuse large B-cell lymphoma; BLL/L, B lymphoblastic lymphoma/leukemia; PTCL-U, peripheral T cell lymphoma, unspecified; ALCL, anaplastic large cell lymphoma; N-NK/T, nasal type NK/T cell lymphoma; AITL, angioimmunoblastic T-cell lymphoma; ATL/L, adult T cell leukemia/lymphoma; TLL/L, T lymphoblastic leukemia/lymphoma.
Values are presented as number (%). IG, immunoglobulin; TCR, T cell receptor; BCL, B cell lymphoma; BLL/L, B lymphoblastic lymphoma/leukemia; TCL, T cell lymphoma; TLL/L, T lymphoblastic leukemia/lymphoma; RLD, Reactive lymphoproliferative disease.
Values are presented as number (%). MCL, mantle cell lymphoma; FL, follicular lymphoma; MZL, marginal zone lymphoma; MALTL, extranodal marginal zone lymphoma of mucosa associated lymphoid tissue; DLBCL, diffuse large B-cell lymphoma; BLL/L, B lymphoblastic leukemia/lymphoma; IG, immunoglobulin; TCR, T cell receptor.
TCL, T cell lymphoma; PTCL-U, peripheral T cell lymphoma, unspecified; ALCL, anaplastic large cell lymphoma; N-NK/T, nasal type NK/T cell lymphoma; AITL, angioimmunoblastic T-cell lymphoma; ATL/L, adult T cell leukemia/lymphoma; TLL/L, T lymphoblastic leukemia/lymphoma; TCR, T cell receptor; IG, immunoglobulin

 E-submission
E-submission