Search
- Page Path
- HOME > Search
Review Article
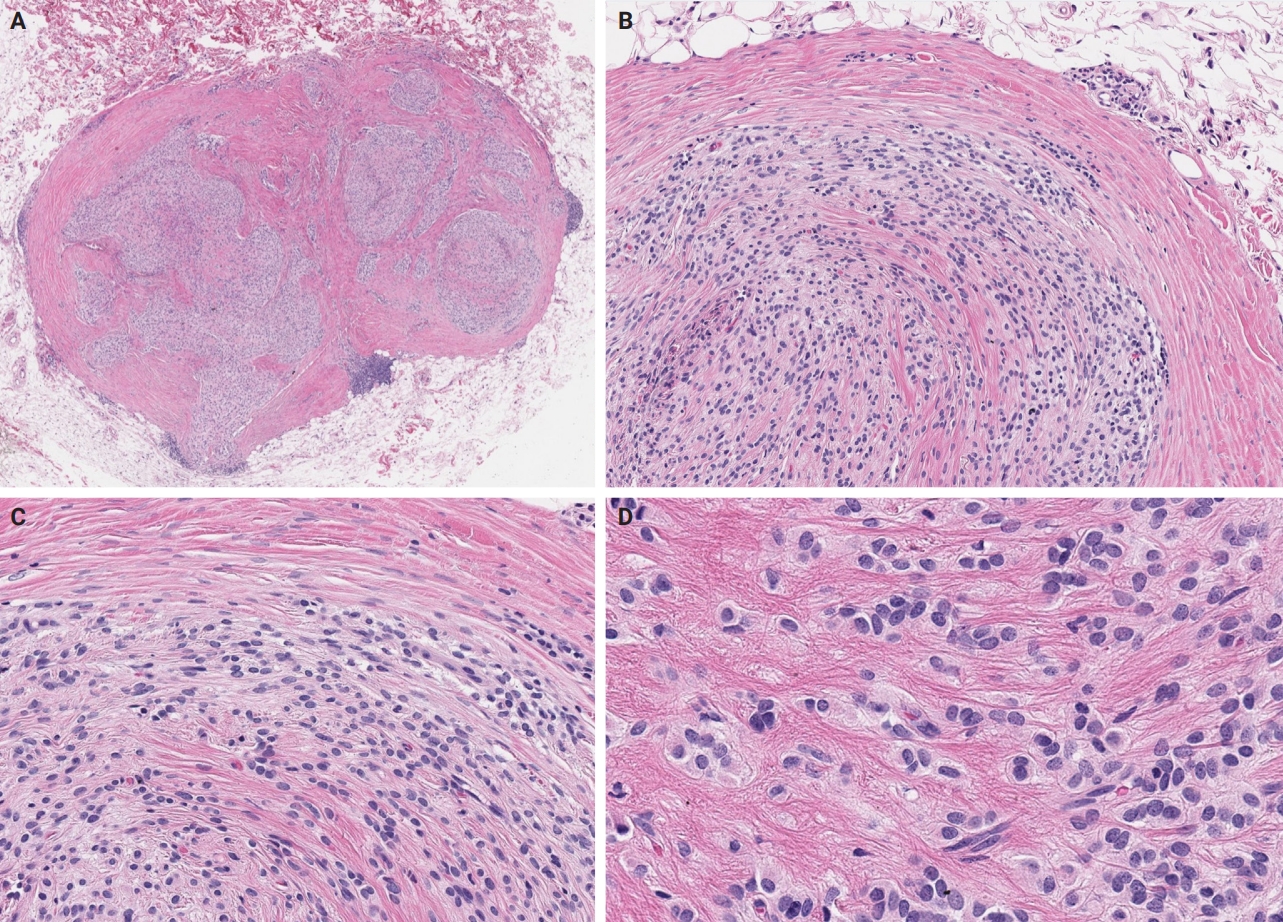
- A comprehensive review of ossifying fibromyxoid tumor: insights into its clinical, pathological, and molecular landscape
- Kyriakos Chatzopoulos, Antonia Syrnioti, Mohamed Yakoub, Konstantinos Linos
- J Pathol Transl Med. 2026;60(1):6-19. Published online January 14, 2026
- DOI: https://doi.org/10.4132/jptm.2025.10.02
- 430 View
- 38 Download
-
 Abstract
Abstract
 PDF
PDF - Ossifying fibromyxoid tumor (OFMT) is a rare mesenchymal neoplasm first described in 1989. It typically arises in the superficial soft tissues of the extremities as a slow-growing, painless mass. Histologically, it is commonly characterized by a multilobular architecture composed of uniform epithelioid cells embedded in a fibromyxoid matrix, often surrounded by a rim of metaplastic bone. While classic cases are readily identifiable, the tumor's histopathological heterogeneity can mimic a range of benign and malignant neoplasms, posing significant diagnostic challenges. Molecularly, most OFMTs harbor PHF1 rearrangements, commonly involving fusion partners such as EP400, MEAF6, or TFE3. This review underscores the importance of an integrated diagnostic approach- incorporating histopathological, immunohistochemical, and molecular data- to accurately classify OFMT and distinguish it from its mimics. Expanding awareness of its morphologic and molecular spectrum is essential for precise diagnosis, optimal patient management, and a deeper understanding of this enigmatic neoplasm.
Review
- Next step of molecular pathology: next-generation sequencing in cytology
- Ricella Souza da Silva, Fernando Schmitt
- J Pathol Transl Med. 2024;58(6):291-298. Published online November 7, 2024
- DOI: https://doi.org/10.4132/jptm.2024.10.22
- 6,000 View
- 366 Download
- 3 Web of Science
- 3 Crossref
-
 Abstract
Abstract
 PDF
PDF - The evolving landscape of precision oncology underscores the pivotal shift from morphological diagnosis to treatment decisions driven by molecular profiling. Recent guidelines from the European Society for Medical Oncology recomend the use of next-generation sequencing (NGS) across a broader range of cancers, reflecting its superior efficiency and clinical value. NGS not only updates oncology testing by offering quicker, sample-friendly, and sensitive analysis but also reduces the need for multiple individual tests. Cytology samples, often obtained through less invasive methods, can yield high-quality genetic material suitable for molecular analysis. This article focuses on optimizing the use of cytology samples in NGS, and outlines their potential benefits in identifying actionable molecular alterations for targeted therapies across various solid tumors. It also addresses the need for validation studies and the strategies to incorporate or combine different types of samples into routine clinical practice. Integrating cytological and liquid biopsies into routine clinical practice, alongside conventional tissue biopsies, offers a comprehensive approach to tumor genotyping, early disease detection, and monitoring of therapeutic responses across various solid tumor types. For comprehensive biomarker characterization, all patient specimens, although limited, is always valuable.
-
Citations
Citations to this article as recorded by- The World Health Organization Reporting System for Lymph Node, Spleen, and Thymus Cytopathology: Part 1 – Lymph Node
Immacolata Cozzolino, Mats Ehinger, Maria Calaminici, Andrea Ronchi, Mousa A. Al-Abbadi, Helena Barroca, Beata Bode-Lesniewska, David F. Chhieng, Ruth L. Katz, Oscar Lin, L. Jeffrey Medeiros, Martha Bishop Pitman, Arvind Rajwanshi, Fernando C. Schmitt, Ph
Acta Cytologica.2025; : 1. CrossRef - The impact of cytological preparation techniques on RNA quality: A comparative study on smear samples
Cisel Aydin Mericoz, Gulsum Caylak, Elif Sevin Sanioglu, Zeynep Seçil Satilmis, Ayse Humeyra Dur Karasayar, Ibrahim Kulac
Cancer Cytopathology.2025;[Epub] CrossRef - Reimagining cytopathology in the molecular era: Integration or fragmentation?
Sumanta Das, R. Naveen Kumar, Biswajit Dey, Pranjal Kalita
Cytojournal.2025; 22: 94. CrossRef
- The World Health Organization Reporting System for Lymph Node, Spleen, and Thymus Cytopathology: Part 1 – Lymph Node
Original Articles
- Establishing molecular pathology curriculum for pathology trainees and continued medical education: a collaborative work from the Molecular Pathology Study Group of the Korean Society of Pathologists
- Jiwon Koh, Ha Young Park, Jeong Mo Bae, Jun Kang, Uiju Cho, Seung Eun Lee, Haeyoun Kang, Min Eui Hong, Jae Kyung Won, Youn-La Choi, Wan-Seop Kim, Ahwon Lee
- J Pathol Transl Med. 2023;57(5):265-272. Published online September 15, 2023
- DOI: https://doi.org/10.4132/jptm.2023.08.26
- 5,359 View
- 207 Download
- 1 Web of Science
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF - Background
The importance of molecular pathology tests has increased during the last decade, and there is a great need for efficient training of molecular pathology for pathology trainees and as continued medical education.
Methods
The Molecular Pathology Study Group of the Korean Society of Pathologists appointed a task force composed of experienced molecular pathologists to develop a refined educational curriculum of molecular pathology. A 3-day online educational session was held based on the newly established structure of learning objectives; the audience were asked to score their understanding of 22 selected learning objectives before and after the session to assess the effect of structured education.
Results
The structured objectives and goals of molecular pathology was established and posted as a web-based interface which can serve as a knowledge bank of molecular pathology. A total of 201 pathologists participated in the educational session. For all 22 learning objectives, the scores of self-reported understanding increased after educational session by 9.9 points on average (range, 6.6 to 17.0). The most effectively improved items were objectives from next-generation sequencing (NGS) section: ‘NGS library preparation and quality control’ (score increased from 51.8 to 68.8), ‘NGS interpretation of variants and reference database’ (score increased from 54.1 to 68.0), and ‘whole genome, whole exome, and targeted gene sequencing’ (score increased from 58.2 to 71.2). Qualitative responses regarding the adequacy of refined educational curriculum were collected, where favorable comments dominated.
Conclusions
Approach toward the education of molecular pathology was refined, which would greatly benefit the future trainees. -
Citations
Citations to this article as recorded by- Presence of RB1 or Absence of LRP1B Mutation Predicts Poor Overall Survival in Patients with Gastric Neuroendocrine Carcinoma and Mixed Adenoneuroendocrine Carcinoma
In Hye Song, Bokyung Ahn, Young Soo Park, Deok Hoon Kim, Seung-Mo Hong
Cancer Research and Treatment.2025; 57(2): 492. CrossRef
- Presence of RB1 or Absence of LRP1B Mutation Predicts Poor Overall Survival in Patients with Gastric Neuroendocrine Carcinoma and Mixed Adenoneuroendocrine Carcinoma
- Single-center study on clinicopathological and typical molecular pathologic features of metastatic brain tumor
- Su Hwa Kim, Young Suk Lee, Sung Hak Lee, Yeoun Eun Sung, Ahwon Lee, Jun Kang, Jae-Sung Park, Sin Soo Jeun, Youn Soo Lee
- J Pathol Transl Med. 2023;57(4):217-231. Published online July 11, 2023
- DOI: https://doi.org/10.4132/jptm.2023.06.10
- 5,330 View
- 173 Download
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF - Background
The metastatic brain tumor is the most common brain tumor. The aim of this study was to demonstrate the clinicopathological and molecular pathologic features of brain metastases (BM).
Methods
A total of 269 patients were diagnosed with BM through surgical resection at Seoul St. Mary’s Hospital from January 2010 to March 2020. We reviewed the clinicopathological features and molecular status of primary and metastatic brain tissues using immunohistochemistry and molecular pathology results.
Results
Among 269 patients, 139 males and 130 females were included. The median age of primary tumor was 58 years (range, 13 to 87 years) and 86 patients (32.0%) had BM at initial presentation. Median BM free interval was 28.0 months (range, 1 to 286 months). The most frequent primary site was lung 46.5% (125/269), and followed by breast 15.6% (42/269), colorectum 10.0% (27/269). Epidermal growth factor receptor (EGFR) mutation was found in 50.8% (32/63) and 58.0% (40/69) of lung primary and BM, respectively. In both breast primary and breast cancer with BM, luminal B was the most frequent subtype at 37.9% (11/29) and 42.9% (18/42), respectively, followed by human epidermal growth factor receptor 2 with 31.0% (9/29) and 33.3% (14/42). Triple-negative was 20.7% (6/29) and 16.7% (7/42), and luminal A was 10.3% (3/29) and 7.1% (3/42) of breast primary and BM, respectively. In colorectal primary and colorectal cancer with BM, KRAS mutation was found in 76.9% (10/13) and 66.7% (2/3), respectively.
Conclusions
We report the clinicopathological and molecular pathologic features of BM that can provide useful information for understanding the pathogenesis of metastasis and for clinical trials based on the tumor’s molecular pathology. -
Citations
Citations to this article as recorded by- Colorectal cancer metastasis to the brain: A scoping review of incidence, treatment, and outcomes
Hunter J Hutchinson, Melanie Gonzalez, Diana Feier, Colin E Welch, Brandon Lucke-Wold
World Journal of Gastrointestinal Pathophysiology.2025;[Epub] CrossRef
- Colorectal cancer metastasis to the brain: A scoping review of incidence, treatment, and outcomes
Reviews
- A standardized pathology report for gastric cancer: 2nd edition
- Young Soo Park, Myeong-Cherl Kook, Baek-hui Kim, Hye Seung Lee, Dong-Wook Kang, Mi-Jin Gu, Ok Ran Shin, Younghee Choi, Wonae Lee, Hyunki Kim, In Hye Song, Kyoung-Mee Kim, Hee Sung Kim, Guhyun Kang, Do Youn Park, So-Young Jin, Joon Mee Kim, Yoon Jung Choi, Hee Kyung Chang, Soomin Ahn, Mee Soo Chang, Song-Hee Han, Yoonjin Kwak, An Na Seo, Sung Hak Lee, Mee-Yon Cho
- J Pathol Transl Med. 2023;57(1):1-27. Published online January 15, 2023
- DOI: https://doi.org/10.4132/jptm.2022.12.23
- 34,199 View
- 1,526 Download
- 22 Web of Science
- 19 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - The first edition of ‘A Standardized Pathology Report for Gastric Cancer’ was initiated by the Gastrointestinal Pathology Study Group of the Korean Society of Pathologists and published 17 years ago. Since then, significant advances have been made in the pathologic diagnosis, molecular genetics, and management of gastric cancer (GC). To reflect those changes, a committee for publishing a second edition of the report was formed within the Gastrointestinal Pathology Study Group of the Korean Society of Pathologists. This second edition consists of two parts: standard data elements and conditional data elements. The standard data elements contain the basic pathologic findings and items necessary to predict the prognosis of GC patients, and they are adequate for routine surgical pathology service. Other diagnostic and prognostic factors relevant to adjuvant therapy, including molecular biomarkers, are classified as conditional data elements to allow each pathologist to selectively choose items appropriate to the environment in their institution. We trust that the standardized pathology report will be helpful for GC diagnosis and facilitate large-scale multidisciplinary collaborative studies.
-
Citations
Citations to this article as recorded by- Spatial and Temporal Tumor Heterogeneity in Gastric Cancer: Discordance of Predictive Biomarkers
Hye Seung Lee
Journal of Gastric Cancer.2025; 25(1): 192. CrossRef - PD-L1 as a Biomarker in Gastric Cancer Immunotherapy
Yunjoo Cho, Soomin Ahn, Kyoung-Mee Kim
Journal of Gastric Cancer.2025; 25(1): 177. CrossRef - Korean Gastric Cancer Association-Led Nationwide Survey on Surgically Treated Gastric Cancers in 2023
Dong Jin Kim, Jeong Ho Song, Ji-Hyeon Park, Sojung Kim, Sin Hye Park, Cheol Min Shin, Yoonjin Kwak, Kyunghye Bang, Chung-sik Gong, Sung Eun Oh, Yoo Min Kim, Young Suk Park, Jeesun Kim, Ji Eun Jung, Mi Ran Jung, Bang Wool Eom, Ki Bum Park, Jae Hun Chung, S
Journal of Gastric Cancer.2025; 25(1): 115. CrossRef - A Comprehensive and Comparative Review of Global Gastric Cancer Treatment Guidelines: 2024 Update
Sang Soo Eom, Keun Won Ryu, Hye Sook Han, Seong-Ho Kong
Journal of Gastric Cancer.2025; 25(1): 153. CrossRef - Korea, Japan, Europe, and the United States: Why are guidelines for gastric cancer different?
Emily E. Stroobant, Seong-Ho Kong, Maria Bencivenga, Takahiro Kinoshita, Tae-Han Kim, Takeshi Sano, Giovanni de Manzoni, Han-Kwang Yang, Yuko Kitagawa, Vivian E. Strong
Gastric Cancer.2025; 28(4): 559. CrossRef - Can the Japanese guidelines for endoscopic submucosal dissection be safely applied to Korean gastric cancer patients? A multicenter retrospective study based on the Korean Gastric Cancer Association nationwide survey
Hayemin Lee, Mi Ryeong Park, Junhyun Lee
Annals of Surgical Treatment and Research.2025; 109(2): 81. CrossRef - Double optimal transport for differential gene regulatory network inference with unpaired samples
Mengyu Li, Bencong Zhu, Cheng Meng, Xiaodan Fan, Laura Cantini
Bioinformatics.2025;[Epub] CrossRef - A Randomized Controlled Trial to Evaluate the Effect of Fibrin Glue on Bleeding after Gastric Endoscopic Submucosal Dissection
Tae-Se Kim, Tae-Jun Kim, Yang Won Min, Hyuk Lee, Byung-Hoon Min, Jun Haeng Lee, Poong-Lyul Rhee, Jae J. Kim
Gut and Liver.2025; 19(5): 677. CrossRef - Diagnostic accuracy of stereomicroscopy assessment of invasion depth in ex vivo specimens of early gastric cancer
Jing Wang, Lin Chang, Dong-Feng Niu, Yan Yan, Chang-Qi Cao, Shi-Jie Li, Qi Wu
World Journal of Gastroenterology.2025;[Epub] CrossRef - SMMILe enables accurate spatial quantification in digital pathology using multiple-instance learning
Zeyu Gao, Anyu Mao, Yuxing Dong, Hannah Clayton, Jialun Wu, Jiashuai Liu, ChunBao Wang, Kai He, Tieliang Gong, Chen Li, Mireia Crispin-Ortuzar
Nature Cancer.2025; 6(12): 2025. CrossRef - Genomic and Transcriptomic Characterization of Gastric Cancer with Bone Metastasis
Sujin Oh, Soo Kyung Nam, Keun-Wook Lee, Hye Seung Lee, Yujun Park, Yoonjin Kwak, Kyu Sang Lee, Ji-Won Kim, Jin Won Kim, Minsu Kang, Young Suk Park, Sang-Hoon Ahn, Yun-Suhk Suh, Do Joong Park, Hyung Ho Kim
Cancer Research and Treatment.2024; 56(1): 219. CrossRef - Microscopic tumor mapping of post-neoadjuvant therapy pancreatic cancer specimens to predict post-surgical recurrence: A prospective cohort study
Yeshong Park, Yeon Bi Han, Jinju Kim, MeeYoung Kang, Boram Lee, Eun Sung Ahn, Saemi Han, Haeryoung Kim, Hee-Young Na, Ho-Seong Han, Yoo-Seok Yoon
Pancreatology.2024; 24(4): 562. CrossRef - Effect of Neoadjuvant Chemotherapy on Tumor-Infiltrating Lymphocytes in Resectable Gastric Cancer: Analysis from a Western Academic Center
Elliott J. Yee, Danielle Gilbert, Jeffrey Kaplan, Sachin Wani, Sunnie S. Kim, Martin D. McCarter, Camille L. Stewart
Cancers.2024; 16(7): 1428. CrossRef - Interpretation of PD-L1 expression in gastric cancer: summary of a consensus meeting of Korean gastrointestinal pathologists
Soomin Ahn, Yoonjin Kwak, Gui Young Kwon, Kyoung-Mee Kim, Moonsik Kim, Hyunki Kim, Young Soo Park, Hyeon Jeong Oh, Kyoungyul Lee, Sung Hak Lee, Hye Seung Lee
Journal of Pathology and Translational Medicine.2024; 58(3): 103. CrossRef - Expression of claudin 18.2 in poorly cohesive carcinoma and its association with clinicopathologic parameters in East Asian patients
Moonsik Kim, Byung Woog Kang, Jihyun Park, Jin Ho Baek, Jong Gwang Kim
Pathology - Research and Practice.2024; 263: 155628. CrossRef - Clinicopathological analysis of claudin 18.2 focusing on intratumoral heterogeneity and survival in patients with metastatic or unresectable gastric cancer
T.-Y. Kim, Y. Kwak, S.K. Nam, D. Han, D.-Y. Oh, S.-A. Im, H.S. Lee
ESMO Open.2024; 9(12): 104000. CrossRef - Pathological Interpretation of Gastric Tumors in Endoscopic Submucosal Dissection
Jung Yeon Kim
Journal of Digestive Cancer Research.2023; 11(1): 15. CrossRef - Histopathology of Gastric Cancer
Baek-hui Kim, Sung Hak Lee
The Korean Journal of Helicobacter and Upper Gastrointestinal Research.2023; 23(2): 143. CrossRef - Endoscopic submucosal dissection hands-on training with artificial mucosal layer EndoGEL
Tae-Se Kim, Jun Haeng Lee
Journal of Innovative Medical Technology.2023; 1(1): 5. CrossRef
- Spatial and Temporal Tumor Heterogeneity in Gastric Cancer: Discordance of Predictive Biomarkers
- Good Laboratory Standards for Clinical Next-Generation Sequencing Cancer Panel Tests
- Jihun Kim, Woong-Yang Park, Nayoung K. D. Kim, Se Jin Jang, Sung-Min Chun, Chang-Ohk Sung, Jene Choi, Young-Hyeh Ko, Yoon-La Choi, Hyo Sup Shim, Jae-Kyung Won
- J Pathol Transl Med. 2017;51(3):191-204. Published online May 10, 2017
- DOI: https://doi.org/10.4132/jptm.2017.03.14
- 29,114 View
- 1,118 Download
- 36 Web of Science
- 37 Crossref
-
 Abstract
Abstract
 PDF
PDF - Next-generation sequencing (NGS) has recently emerged as an essential component of personalized cancer medicine due to its high throughput and low per-base cost. However, no sufficient guidelines for implementing NGS as a clinical molecular pathology test are established in Korea. To ensure clinical grade quality without inhibiting adoption of NGS, a taskforce team assembled by the Korean Society of Pathologists developed laboratory guidelines for NGS cancer panel testing procedures and requirements for clinical implementation of NGS. This consensus standard proposal consists of two parts: laboratory guidelines and requirements for clinical NGS laboratories. The laboratory guidelines part addressed several important issues across multistep NGS cancer panel tests including choice of gene panel and platform, sample handling, nucleic acid management, sample identity tracking, library preparation, sequencing, analysis and reporting. Requirements for clinical NGS tests were summarized in terms of documentation, validation, quality management, and other required written policies. Together with appropriate pathologist training and international laboratory standards, these laboratory standards would help molecular pathology laboratories to successfully implement NGS cancer panel tests in clinic. In this way, the oncology community would be able to help patients to benefit more from personalized cancer medicine.
-
Citations
Citations to this article as recorded by- Tumour purity assessment with deep learning in colorectal cancer and impact on molecular analysis
Lydia A Schoenpflug, Aikaterini Chatzipli, Korsuk Sirinukunwattana, Susan Richman, Andrew Blake, James Robineau, Kirsten D Mertz, Clare Verrill, Simon J Leedham, Claire Hardy, Celina Whalley, Keara Redmond, Philip Dunne, Steven Walker, Andrew D Beggs, Ult
The Journal of Pathology.2025; 265(2): 184. CrossRef - Clinical Validation of Local Versus Commercial Genomic Testing in Cancer: A Comparison of Tissue and Plasma Concordance
Lucy G. Faulkner, Lynne Howells, Susann Lehman, Caroline Cowley, Zahirah Sidat, Jacqui Shaw, Anne L. Thomas
Cancer Investigation.2025; 43(2): 119. CrossRef - Diagnostic Implications of NGS-Based Molecular Profiling in Mature B-Cell Lymphomas with Potential Bone Marrow Involvement
Bernhard Strasser, Sebastian Mustafa, Josef Seier, Erich Wimmer, Josef Tomasits
Diagnostics.2025; 15(6): 727. CrossRef - Pragmatic nationwide master observational trial based on genomic alterations in advanced solid tumors: KOrean Precision Medicine Networking Group Study of MOlecular profiling guided therapy based on genomic alterations in advanced Solid tumors (KOSMOS)-II
Sun Young Kim, Jee Hyun Kim, Tae-Yong Kim, Sook Ryun Park, Shinkyo Yoon, Soohyeon Lee, Se-Hoon Lee, Tae Min Kim, Sae-Won Han, Hye Ryun Kim, Hongseok Yun, Sejoon Lee, Jihun Kim, Yoon-La Choi, Kui Son Choi, Heejung Chae, Hyewon Ryu, Gyeong-Won Lee, Dae Youn
BMC Cancer.2024;[Epub] CrossRef - Reporting of somatic variants in clinical cancer care: recommendations of the Swiss Society of Molecular Pathology
Yann Christinat, Baptiste Hamelin, Ilaria Alborelli, Paolo Angelino, Valérie Barbié, Bettina Bisig, Heather Dawson, Milo Frattini, Tobias Grob, Wolfram Jochum, Ronny Nienhold, Thomas McKee, Matthias Matter, Edoardo Missiaglia, Francesca Molinari, Sacha Ro
Virchows Archiv.2024; 485(6): 1033. CrossRef - Acute myeloid leukemia and myelodysplastic neoplasms: clinical implications of myelodysplasia-related genes mutations and TP53 aberrations
Hyunwoo Kim, Ja Young Lee, Shinae Yu, Eunkyoung Yoo, Hye Ran Kim, Sang Min Lee, Won Sik Lee
Blood Research.2024;[Epub] CrossRef - Validation and Clinical Application of ONCOaccuPanel for Targeted Next-Generation Sequencing of Solid Tumors
Moonsik Kim, Changseon Lee, Juyeon Hong, Juhee Kim, Ji Yun Jeong, Nora Jee-Young Park, Ji-Eun Kim, Ji Young Park
Cancer Research and Treatment.2023; 55(2): 429. CrossRef - Establishing molecular pathology curriculum for pathology trainees and continued medical education: a collaborative work from the Molecular Pathology Study Group of the Korean Society of Pathologists
Jiwon Koh, Ha Young Park, Jeong Mo Bae, Jun Kang, Uiju Cho, Seung Eun Lee, Haeyoun Kang, Min Eui Hong, Jae Kyung Won, Youn-La Choi, Wan-Seop Kim, Ahwon Lee
Journal of Pathology and Translational Medicine.2023; 57(5): 265. CrossRef - Clinical applications of next-generation sequencing in the diagnosis of genetic disorders in Korea: a narrative review
Jihoon G. Yoon, Man Jin Kim, Yong Jin Kwon, Jong-Hee Chae
Journal of the Korean Medical Association.2023; 66(10): 613. CrossRef - Obtaining spatially resolved tumor purity maps using deep multiple instance learning in a pan-cancer study
Mustafa Umit Oner, Jianbin Chen, Egor Revkov, Anne James, Seow Ye Heng, Arife Neslihan Kaya, Jacob Josiah Santiago Alvarez, Angela Takano, Xin Min Cheng, Tony Kiat Hon Lim, Daniel Shao Weng Tan, Weiwei Zhai, Anders Jacobsen Skanderup, Wing-Kin Sung, Hwee
Patterns.2022; 3(2): 100399. CrossRef - Update on Molecular Diagnosis in Extranodal NK/T-Cell Lymphoma and Its Role in the Era of Personalized Medicine
Ka-Hei (Murphy) Sun, Yin-Ting (Heylie) Wong, Ka-Man (Carmen) Cheung, Carmen (Michelle) Yuen, Yun-Tat (Ted) Chan, Wing-Yan (Jennifer) Lai, Chun (David) Chao, Wing-Sum (Katie) Fan, Yuen-Kiu (Karen) Chow, Man-Fai Law, Ho-Chi (Tommy) Tam
Diagnostics.2022; 12(2): 409. CrossRef - Defining Novel DNA Virus-Tumor Associations and Genomic Correlates Using Prospective Clinical Tumor/Normal Matched Sequencing Data
Chad M. Vanderbilt, Anita S. Bowman, Sumit Middha, Kseniya Petrova-Drus, Yi-Wei Tang, Xin Chen, Youxiang Wang, Jason Chang, Natasha Rekhtman, Klaus J. Busam, Sounak Gupta, Meera Hameed, Maria E. Arcila, Marc Ladanyi, Michael F. Berger, Snjezana Dogan, Ahm
The Journal of Molecular Diagnostics.2022; 24(5): 515. CrossRef - Performance Evaluation of Three DNA Sample Tracking Tools in a Whole Exome Sequencing Workflow
Gertjan Wils, Céline Helsmoortel, Pieter-Jan Volders, Inge Vereecke, Mauro Milazzo, Jo Vandesompele, Frauke Coppieters, Kim De Leeneer, Steve Lefever
Molecular Diagnosis & Therapy.2022; 26(4): 411. CrossRef - Clinical Quality Considerations when Using Next-Generation Sequencing (NGS) in Clinical Drug Development
Timothé Ménard, Alaina Barros, Christopher Ganter
Therapeutic Innovation & Regulatory Science.2021; 55(5): 1066. CrossRef - Fast Healthcare Interoperability Resources (FHIR)–Based Quality Information Exchange for Clinical Next-Generation Sequencing Genomic Testing: Implementation Study
Donghyeong Seong, Sungwon Jung, Sungchul Bae, Jongsuk Chung, Dae-Soon Son, Byoung-Kee Yi
Journal of Medical Internet Research.2021; 23(4): e26261. CrossRef - Status of Next-Generation Sequencing-Based Genetic Diagnosis in Hematologic Malignancies in Korea (2017-2018)
JinJu Kim, Ja Young Lee, Jungwon Huh, Myung-Hyun Nam, Myungshin Kim, Young-Uk Cho, Sun-Young Kong, Seung-Tae Lee, In-Suk Kim
Laboratory Medicine Online.2021; 11(1): 25. CrossRef - MSI-Testung
Josef Rüschoff, Gustavo Baretton, Hendrik Bläker, Wolfgang Dietmaier, Manfred Dietel, Arndt Hartmann, Lars-Christian Horn, Korinna Jöhrens, Thomas Kirchner, Ruth Knüchel, Doris Mayr, Sabine Merkelbach-Bruse, Hans-Ulrich Schildhaus, Peter Schirmacher, Mark
Der Pathologe.2021; 42(4): 414. CrossRef - Molecular biomarker testing for non–small cell lung cancer: consensus statement of the Korean Cardiopulmonary Pathology Study Group
Sunhee Chang, Hyo Sup Shim, Tae Jung Kim, Yoon-La Choi, Wan Seop Kim, Dong Hoon Shin, Lucia Kim, Heae Surng Park, Geon Kook Lee, Chang Hun Lee
Journal of Pathology and Translational Medicine.2021; 55(3): 181. CrossRef - MSI testing
Josef Rüschoff, Gustavo Baretton, Hendrik Bläker, Wolfgang Dietmaier, Manfred Dietel, Arndt Hartmann, Lars-Christian Horn, Korinna Jöhrens, Thomas Kirchner, Ruth Knüchel, Doris Mayr, Sabine Merkelbach-Bruse, Hans-Ulrich Schildhaus, Peter Schirmacher, Mark
Der Pathologe.2021; 42(S1): 110. CrossRef - 16S rDNA microbiome composition pattern analysis as a diagnostic biomarker for biliary tract cancer
Huisong Lee, Hyeon Kook Lee, Seog Ki Min, Won Hee Lee
World Journal of Surgical Oncology.2020;[Epub] CrossRef - Risk Stratification Using a Novel Genetic Classifier IncludingPLEKHS1Promoter Mutations for Differentiated Thyroid Cancer with Distant Metastasis
Chan Kwon Jung, Seung-Hyun Jung, Sora Jeon, Young Mun Jeong, Yourha Kim, Sohee Lee, Ja-Seong Bae, Yeun-Jun Chung
Thyroid.2020; 30(11): 1589. CrossRef - Biomarker testing for advanced lung cancer by next-generation sequencing; a valid method to achieve a comprehensive glimpse at mutational landscape
Anurag Mehta, Smreti Vasudevan, Sanjeev Kumar Sharma, Manoj Panigrahi, Moushumi Suryavanshi, Mumtaz Saifi, Ullas Batra
Applied Cancer Research.2020;[Epub] CrossRef - Application Areas of Traditional Molecular Genetic Methods and NGS in relation to Hereditary Urological Cancer Diagnosis
Dmitry S. Mikhaylenko, Alexander S. Tanas, Dmitry V. Zaletaev, Marina V. Nemtsova
Journal of Oncology.2020; 2020: 1. CrossRef - Assembling and Validating Bioinformatic Pipelines for Next-Generation Sequencing Clinical Assays
Jeffrey A SoRelle, Megan Wachsmann, Brandi L. Cantarel
Archives of Pathology & Laboratory Medicine.2020; 144(9): 1118. CrossRef - Standard operating procedure for somatic variant refinement of sequencing data with paired tumor and normal samples
Erica K. Barnell, Peter Ronning, Katie M. Campbell, Kilannin Krysiak, Benjamin J. Ainscough, Lana M. Sheta, Shahil P. Pema, Alina D. Schmidt, Megan Richters, Kelsy C. Cotto, Arpad M. Danos, Cody Ramirez, Zachary L. Skidmore, Nicholas C. Spies, Jasreet Hun
Genetics in Medicine.2019; 21(4): 972. CrossRef - A DNA pool of FLT3-ITD positive DNA samples can be used efficiently for analytical evaluation of NGS-based FLT3-ITD quantitation - Testing several different ITD sequences and rates, simultaneously
Zoltán A. Mezei, Dávid Tornai, Róza Földesi, László Madar, Andrea Sümegi, Mária Papp, Péter Antal-Szalmás
Journal of Biotechnology.2019; 303: 25. CrossRef - Pharmacogenomic Testing: Clinical Evidence and Implementation Challenges
Catriona Hippman, Corey Nislow
Journal of Personalized Medicine.2019; 9(3): 40. CrossRef - Cancer Panel Assay for Precision Oncology Clinic: Results from a 1-Year Study
Dohee Kwon, Binnari Kim, Hyeong Chan Shin, Eun Ji Kim, Sang Yun Ha, Kee-Taek Jang, Seung Tae Kim, Jeeyun Lee, Won Ki Kang, Joon Oh Park, Kyoung-Mee Kim
Translational Oncology.2019; 12(11): 1488. CrossRef - Analytical Evaluation of an NGS Testing Method for Routine Molecular Diagnostics on Melanoma Formalin-Fixed, Paraffin-Embedded Tumor-Derived DNA
Irene Mancini, Lisa Simi, Francesca Salvianti, Francesca Castiglione, Gemma Sonnati, Pamela Pinzani
Diagnostics.2019; 9(3): 117. CrossRef - Benchmark Database for Process Optimization and Quality Control of Clinical Cancer Panel Sequencing
Donghyeong Seong, Jongsuk Chung, Ki-Wook Lee, Sook-Young Kim, Byung-Suk Kim, Jung-Keun Song, Sungwon Jung, Taeseob Lee, Donghyun Park, Byoung-Kee Yi, Woong-Yang Park, Dae-Soon Son
Biotechnology and Bioprocess Engineering.2019; 24(5): 793. CrossRef - Use of the Ion PGM and the GeneReader NGS Systems in Daily Routine Practice for Advanced Lung Adenocarcinoma Patients: A Practical Point of View Reporting a Comparative Study and Assessment of 90 Patients
Simon Heeke, Véronique Hofman, Elodie Long-Mira, Virginie Lespinet, Salomé Lalvée, Olivier Bordone, Camille Ribeyre, Virginie Tanga, Jonathan Benzaquen, Sylvie Leroy, Charlotte Cohen, Jérôme Mouroux, Charles Marquette, Marius Ilié, Paul Hofman
Cancers.2018; 10(4): 88. CrossRef - Use of the Ion AmpliSeq Cancer Hotspot Panel in clinical molecular pathology laboratories for analysis of solid tumours: With emphasis on validation with relevant single molecular pathology tests and the Oncomine Focus Assay
Ahwon Lee, Sung-Hak Lee, Chan Kwon Jung, Gyungsin Park, Kyo Young Lee, Hyun Joo Choi, Ki Ouk Min, Tae Jung Kim, Eun Jung Lee, Youn Soo Lee
Pathology - Research and Practice.2018; 214(5): 713. CrossRef - Recent Advancement of the Molecular Diagnosis in Pediatric Brain Tumor
Jeong-Mo Bae, Jae-Kyung Won, Sung-Hye Park
Journal of Korean Neurosurgical Society.2018; 61(3): 376. CrossRef - The long tail of molecular alterations in non-small cell lung cancer: a single-institution experience of next-generation sequencing in clinical molecular diagnostics
Caterina Fumagalli, Davide Vacirca, Alessandra Rappa, Antonio Passaro, Juliana Guarize, Paola Rafaniello Raviele, Filippo de Marinis, Lorenzo Spaggiari, Chiara Casadio, Giuseppe Viale, Massimo Barberis, Elena Guerini-Rocco
Journal of Clinical Pathology.2018; 71(9): 767. CrossRef - Clinical laboratory utilization management and improved healthcare performance
Christopher Naugler, Deirdre L. Church
Critical Reviews in Clinical Laboratory Sciences.2018; 55(8): 535. CrossRef - Development of HLA-A, -B and -DR Typing Method Using Next-Generation Sequencing
Dong Hee Seo, Jeong Min Lee, Mi Ok Park, Hyun Ju Lee, Seo Yoon Moon, Mijin Oh, So Young Kim, Sang-Heon Lee, Ki-Eun Hyeong, Hae-Jin Hu, Dae-Yeon Cho
The Korean Journal of Blood Transfusion.2018; 29(3): 310. CrossRef - Value-based genomics
Jun Gong, Kathy Pan, Marwan Fakih, Sumanta Pal, Ravi Salgia
Oncotarget.2018; 9(21): 15792. CrossRef
- Tumour purity assessment with deep learning in colorectal cancer and impact on molecular analysis

 E-submission
E-submission





 First
First Prev
Prev



