Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 56(2); 2022 > Article
-
Newsletter
What’s new in molecular genetic pathology 2022: immune checkpoint inhibitor biomarkers and select solid tumors -
Patricia C. Tsang, MD1
 , Guoli Chen, MD2
, Guoli Chen, MD2
-
Journal of Pathology and Translational Medicine 2022;56(2):113-114.
DOI: https://doi.org/10.4132/jptm.2022.01.25
Published online: March 11, 2022
1MedStar Washington Hospital Center, Pathology & Laboratory Medicine, Washington, DC, USA
2Geisinger Medical Laboratories, Geisinger Health, Danville, Pennsylvania, USA
- Corresponding Author: Patricia C. Tsang, MD MedStar Washington Hospital Center, Pathology & Laboratory Medicine, Washington, DC, USA E-mail: patctsang@hotmail.com
- This article has been published jointly, with consent, in both Journal of Pathology and Translational Medicine and PathologyOutlines.com.
© 2022 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
- Predictive biomarker testing plays a critical role in targeted immuno-oncology, including the use of immune checkpoint inhibitors (ICI) for various solid tumors. Molecular advancements in cancers of the breast, kidney and brain have continued to propel tumor classification and precision therapy.
- The HUGO Gene Nomenclature Committee has released a new recommendation to denote gene fusion with a double colon, e.g., BCR::ABL [1].
GENE FUSION NOMENCLATURE
- • Programmed cell death ligand 1 (PDL1) immunohistochemistry (IHC) uses various scoring systems and cutoffs for particular antibody clones to quantify PDL1 expression as a predictive biomarker for specific FDA approved ICI. For example:
- The PDL1 pharmDX 22C3 immunostain is a companion diagnostic for non small cell lung carcinoma (NSCLC) patients eligible for pembrolizumab immunotherapy.
- The tumor proportion score (TPS) is calculated by dividing the total number of PDL1 positive tumor cells by the total number of tumor cells (≥ 1% positive cutoff).
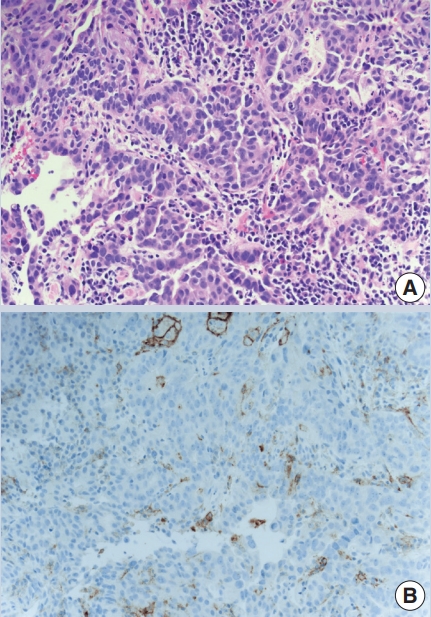
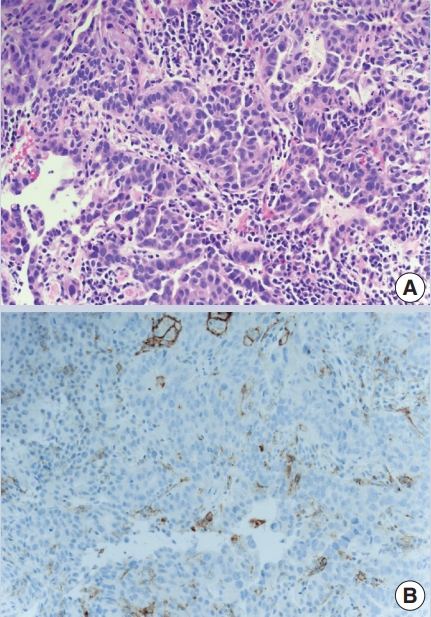
- The combined positive score (CPS) is based on dividing the number of PDL1 positive cells (tumor cells, lymphocytes, macrophages) by the number of tumor cells to predict pembrolizumab efficacy in metastatic gastric and gastroesophageal junctional carcinoma, bladder cancer and cervical cancer (Fig. 1). It has the same cutoff of ≥ 1%, using the 22C3 antibody clone.
- The PDL1 SP142 clone is an FDA approved complementary diagnostic to predict response of metastatic NSCLC to atezolizumab. The evaluation is based on either the proportion of tumor area occupied by PDL1 expressing tumor infiltrating immune cells of any intensity or the percentage of PDL1 expressing tumor cells of any intensity.
- • Deficient mismatch repair (dMMR) / high microsatellite instability (MSI-H) is predictive of ICI efficacy in colorectal cancers and other solid tumors. While it is commonly tested by IHC, assays based on PCR or next generation sequencing (NGS) are available.
- • High tumor mutational burden (TMB) may also serve as a predictor of immunotherapy response. TMB reflects the number of somatic mutations per megabase of tumor genomic sequence. It can be determined by whole exome sequencing (WES) or targeted sequencing of gene panels (usually ≥ 200 genes). Cutoff values have not yet been unified and may vary in respect to different assay platforms and ICI drug targets. Recently, Foundation Medicine’s FoundationOne® CDx, which includes MSI and TMB assessment, was approved as a companion diagnostic for pembrolizumab with a cutoff of ≥ 10 mutations per megabase, regardless of solid tumor type.
IMMUNE CHECKPOINT INHIBITOR BIOMARKERS
- • NGS can simultaneously detect alterations of multiple genes involved in breast cancers. Clinically actionable genomic changes include ERBB2 (HER2) amplification (prevalence ~15%), BRCA1/BRCA2 mutations (5%–10%), PIK3CA mutations (30%–40%) and ETV6::NTRK3 fusion in secretory breast carcinoma (< 0.2% of breast cancers).
- BRCA1 and BRCA2 inactivating mutations carry a lifetime cumulative risk of 60% and 55% to develop breast cancer, respectively. Poly-ADP-ribose polymerase (PARP) inhibitors are FDA approved targeted therapy for BRCA1/BRCA2 associated breast cancer.
- API3K (phosphoinositide 3-kinase) inhibitor, alpelisib, is FDA approved for advanced breast cancers that are PIK3CA altered, HER2 (-) and hormone receptor (+).
- Seen in ~30% of breast cancers, TP53 mutation is associated with an increased likelihood of pathologic complete remission following chemotherapy, while wild type TP53 appears to induce cell cycle arrest instead of cell death, resulting in residual disease following chemotherapy [2].
- • Multigenomic prognostic profiling is being used to predict the risk of breast cancer recurrence. Commercial platforms include:
- Oncotype DX® (Genomic Health Inc.): 21 gene reverse transcription PCR assay for predicting the likelihood of recurrence and chemotherapy benefit in hormone receptor (+) patients on endocrine therapy.
- MammaPrint (Agendia, Inc.): 70 gene expression profile by microarray for predicting the risk of distant metastasis. Patients with high clinical risk but low genomic risk based on MammaPrint might not require chemotherapy.
- EndoPredict® (Myriad Genetics): 11 gene RNA expression profile for predicting the risk of recurrence in hormone receptor (+) / HER2 (-) patients who are node (+) or (-) on endocrine therapy.
- Prosigna® (Veracyte): 50 gene reverse transcription PCR assay for assessing the risk of recurrence in 10 years for hormone receptor (+) early stage breast cancer and identifying the subset of patients who may not benefit from chemotherapy.
BREAST CARCINOMA
- • In addition to recurrent VHL alterations and 3p deletion, clear cell RCC frequently carries other genetic alterations. BAP1 or SETD2 mutated RCCs behave aggressively, while PBRM1 mutations predict a poor prognosis in localized RCC but a favorable outcome in advanced disease.
- • Chromosome 7 or 17 polysomy and trisomy are the most common chromosomal changes in type 1 papillary RCC. Type 2 tumors are heterogeneous and may harbor CDKN2A silencing, SETD2 mutations and other alterations.
- • Some hybrid oncocytic chromophobe tumors (HOCT) are associated with Birt-Hogg-Dubé syndrome (BHD) or renal oncocytosis, while others are sporadic. BHD can be confirmed by germline mutations in the FLCN tumor suppressor gene.
- • Renal cell neoplasms with TFE3, TFEB and MITF rearrangements are now grouped together as MiT family translocation RCC.
- • Loss of SMARCB1 expression by IHC supports a diagnosis of medullary carcinoma.
- • Negative or abnormal IHC staining for fumarate hydratase (FH), positive IHC staining for 2-succino-cysteine (2SC) or FH gene mutation supports FH deficient RCC / hereditary leiomyomatosis and RCC (HLRCC). Succinate dehydrogenase (SDH) deficient RCCs are associated with germline mutations of the SDH subunits (usually SDHB), as demonstrable by negative SDHB IHC staining. Genetic counseling is recommended for FH-deficient or SDH deficient RCC.
- • Molecular profiling has helped to define new and emerging RCC variants, e.g., eosinophilic solid and cystic RCC associated with tuberous sclerosis (TSC1 or TSC2 gene alterations), RCC with TSC/MTOR mutations, TCEB1 mutated RCC, RCC with TFEB/6p21/VEGFA amplification and ALK rearranged RCC [3].
RENAL CELL CARCINOMA (RCC)
- • Primary glioblastoma (GBM) may harbor 10q loss of heterozygosity (LOH) (70%), EGFR amplification (36%), CDKN2A deletion (31%) and PTEN mutations (25%), all of which denote aggressive disease.
- • MGMT promoter hypermethylation (30%–40% of primary GBM) predicts improved response to alkylating agents and a lower risk of tumor recurrence.
- • Associated with improved survival, IDH1 or IDH2 mutations are common in grades II and III astrocytoma, oligodendroglioma and secondary GBM, and uncommon in primary GBM. When present, they help to distinguish low-grade astrocytoma (~70%) from gliosis.
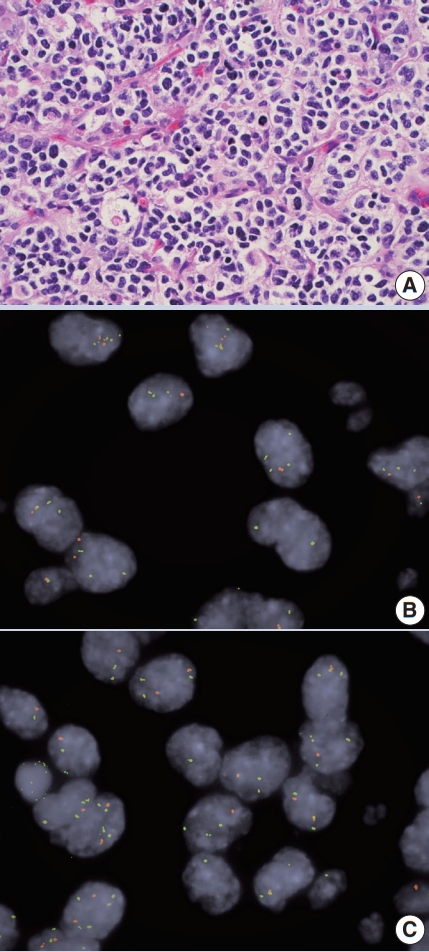
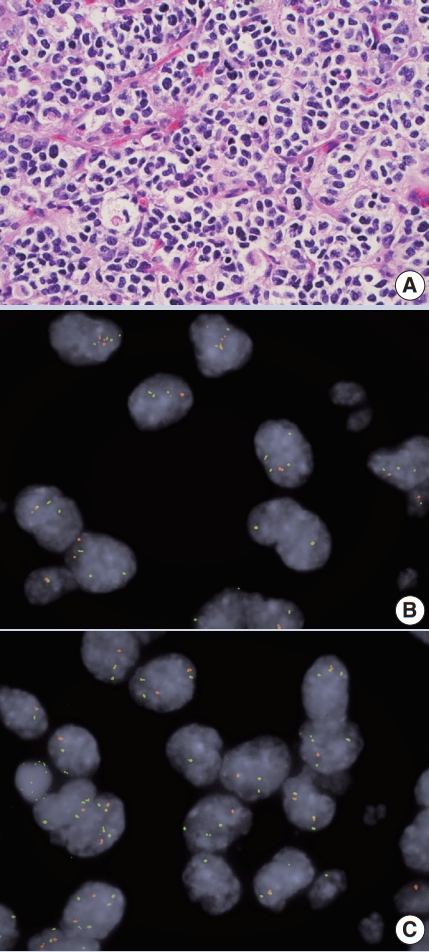
- • Co-deletion of 1p / 19q (detectable by FISH / PCR / microarray) is characteristic of oligodendroglioma (Fig. 2), predicting response to conventional therapy. Equivocal FISH can be subsequently assessed by LOH analysis via PCR. ATRX loss of function mutations, which correlate well with negative IHC, favor diffuse astrocytoma over oligodendroglioma.
- • Diffuse midline glioma is characterized by a K27M substitution in the histone H3 genes, H3F3A or HIST1H3B/HIST1H3C, resulting in hypomethylation and H3 protein inactivation. These tumors may also harbor mutations in TP53 (42%) or ACVR1 (24%) [4].
- • BRAF fusion, most commonly with KIAA1549, is observed in 59%–90% of pilocytic astrocytomas.
BRAIN TUMORS
- Dr. Tsang joined the Pathology Outlines editorial board in 2019 and has been serving as Deputy Editor in Chief for Clinical Pathology for PathologyOutlines since 2020. She works as Chair of Pathology & Laboratory Medical Director at MedStar Washington Hospital Center in Washington, D.C. She has been practicing molecular pathology for more than a decade.
- Dr. Chen is currently an Associate and Attending Pathologist at Geisinger Medical Center, where he participates in the clinical, academic and teaching services in surgical pathology and molecular diagnostics. He has been actively publishing and lecturing on topics related to molecular pathology of solid tumors.
Meet the Authors
- 1. Bruford EA, Antonescu CR, Carroll AJ, et al. HUGO Gene Nomenclature Committee (HGNC) recommendations for the designation of gene fusions. Leukemia 2021; 35: 3040-3. ArticlePubMedPMCPDF
- 2. Shahbandi A, Nguyen HD, Jackson JG. TP53 mutations and outcomes in breast cancer: reading beyond the headlines. Trends Cancer 2020; 6: 98-110. ArticlePubMedPMC
- 3. Henske EP, Cornejo KM, Wu CL. Renal Cell Carcinoma in tuberous sclerosis complex. Genes (Basel) 2021; 12: 1585.ArticlePubMedPMC
- 4. Price G, Bouras A, Hambardzumyan D, Hadjipanay- is CG. Current knowledge on the immune microenvironment and emerging immunotherapies in diffuse midline glioma. EBioMedicine 2021; 69: 103453.ArticlePubMedPMC
REFERENCES
Figure & Data
References
Citations

 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure
- Related articles
-
- What’s new in hematopathology 2025: myeloid neoplasms in the WHO 5th edition and ICC
- What’s new in thyroid pathology 2024: updates from the new WHO classification and Bethesda system
- What’s new in bone and soft tissue pathology 2023: guidelines for molecular testing
- What’s new in neuromuscular pathology 2022: myopathy updates and gene therapies
- What’s new in breast pathology 2022: WHO 5th edition and biomarker updates



 E-submission
E-submission