Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 53(6); 2019 > Article
-
Original Article
Interobserver Reproducibility of PD-L1 Biomarker in Non-small Cell Lung Cancer: A Multi-Institutional Study by 27 Pathologists -
Sunhee Chang,*
 , Hyung Kyu Park1,*
, Hyung Kyu Park1,* , Yoon-La Choi,2
, Yoon-La Choi,2 , Se Jin Jang3
, Se Jin Jang3 , Cardiopulmonary Pathology Study Group of the Korean Society of Pathologists
, Cardiopulmonary Pathology Study Group of the Korean Society of Pathologists -
Journal of Pathology and Translational Medicine 2019;53(6):347-353.
DOI: https://doi.org/10.4132/jptm.2019.09.29
Published online: October 28, 2019
Department of Pathology, Inje University Ilsan Paik Hospital, Goyang, Korea
1Department of Pathology, Konkuk University Medical Center, Konkuk University School of Medicine, Seoul, Korea
2Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
3Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- Corresponding Author: Yoon-La Choi, MD, Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, 81 Irwon-ro, Gangnam-gu, Seoul 06351, Korea Tel: +82-2-3410-2797, Fax: +82-2-3410-6396, E-mail: ylachoi@skku.edu
- *Sunhee Chang and Hyung Kyu Park contributed equally to this work.
© 2019 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Figure & Data
References
Citations

- PD-L1 expression in PitNETs: Correlations with the 2022 WHO classification
Ethan Harel, Ekkehard Hewer, Stefano La Rosa, Jean Philippe Brouland, Nelly Pitteloud, Federico Santoni, Maxime Brunner, Roy Thomas Daniel, Mahmoud Messerer, Giulia Cossu
Brain and Spine.2025; 5: 104171. CrossRef - Relationship Between Short‐Term Outcomes and PD‐L1 Expression Based on Combined Positive Score and Tumor Proportion Score in Recurrent or Metastatic Head and Neck Cancers Treated With Anti‐PD‐1 Antibody Monotherapy
Akihiro Ohara, Taisuke Mori, Mai Itoyama, Kazuki Yokoyama, Shun Yamamoto, Ken Kato, Yoshitaka Honma
Cancer Reports.2025;[Epub] CrossRef - Convergence of evolving artificial intelligence and machine learning techniques in precision oncology
Elena Fountzilas, Tillman Pearce, Mehmet A. Baysal, Abhijit Chakraborty, Apostolia M. Tsimberidou
npj Digital Medicine.2025;[Epub] CrossRef - PD‐L1 Scoring Models for Non‐Small Cell Lung Cancer in China: Current Status, AI‐Assisted Solutions and Future Perspectives
Ziling Huang, Shen Wang, Jiansong Zhou, Haiquan Chen, Yuan Li
Thoracic Cancer.2025;[Epub] CrossRef - Assessing PD‐L1 expression in non‐small cell lung carcinoma: a prospective study of matched fine‐needle aspirates, core biopsies, and resection specimens using alcohol and forming fixatives
Alexander Haragan, Natalie Kipling, Michael Shackcloth, John R Gosney, Michael P Davies, John K Field
The Journal of Pathology: Clinical Research.2025;[Epub] CrossRef - Weakly Supervised Deep Learning Predicts Immunotherapy Response in Solid Tumors Based on PD-L1 Expression
Marta Ligero, Garazi Serna, Omar S.M. El Nahhas, Irene Sansano, Siarhei Mauchanski, Cristina Viaplana, Julien Calderaro, Rodrigo A. Toledo, Rodrigo Dienstmann, Rami S. Vanguri, Jennifer L. Sauter, Francisco Sanchez-Vega, Sohrab P. Shah, Santiago Ramón y C
Cancer Research Communications.2024; 4(1): 92. CrossRef - Concordance of assessments of four PD-L1 immunohistochemical assays in esophageal squamous cell carcinoma (ESCC)
Xinran Wang, Jiankun He, Jinze Li, Chun Wu, Meng Yue, Shuyao Niu, Ying Jia, Zhanli Jia, Lijing Cai, Yueping Liu
Journal of Cancer Research and Clinical Oncology.2024;[Epub] CrossRef - A Deep Learning–Based Assay for Programmed Death Ligand 1 Immunohistochemistry Scoring in Non–Small Cell Lung Carcinoma: Does it Help Pathologists Score?
Hiroaki Ito, Akihiko Yoshizawa, Kazuhiro Terada, Akiyoshi Nakakura, Mariyo Rokutan-Kurata, Tatsuhiko Sugimoto, Kazuya Nishimura, Naoki Nakajima, Shinji Sumiyoshi, Masatsugu Hamaji, Toshi Menju, Hiroshi Date, Satoshi Morita, Ryoma Bise, Hironori Haga
Modern Pathology.2024; 37(6): 100485. CrossRef - Clinical Validation of Artificial Intelligence–Powered PD-L1 Tumor Proportion Score Interpretation for Immune Checkpoint Inhibitor Response Prediction in Non–Small Cell Lung Cancer
Hyojin Kim, Seokhwi Kim, Sangjoon Choi, Changhee Park, Seonwook Park, Sergio Pereira, Minuk Ma, Donggeun Yoo, Kyunghyun Paeng, Wonkyung Jung, Sehhoon Park, Chan-Young Ock, Se-Hoon Lee, Yoon-La Choi, Jin-Haeng Chung
JCO Precision Oncology.2024;[Epub] CrossRef - Reproducibility of c-Met Immunohistochemical Scoring (Clone SP44) for Non–Small Cell Lung Cancer Using Conventional Light Microscopy and Whole Slide Imaging
Christophe Bontoux, Véronique Hofman, Emmanuel Chamorey, Renaud Schiappa, Sandra Lassalle, Elodie Long-Mira, Katia Zahaf, Salomé Lalvée, Julien Fayada, Christelle Bonnetaud, Samantha Goffinet, Marius Ilié, Paul Hofman
American Journal of Surgical Pathology.2024; 48(9): 1072. CrossRef - A preliminary study on the diagnostic performance of the uPath PD-L1 (SP263) artificial intelligence (AI) algorithm in patients with NSCLC treated with PD-1/PD-L1 checkpoint blockade
Alessio Cortellini, Claudia Zampacorta, Michele De Tursi, Lucia R. Grillo, Serena Ricciardi, Emilio Bria, Maurizio Martini, Raffaele Giusti, Marco Filetti, Antonella Dal Mas, Marco Russano, Filippo Gustavo Dall’Olio, Fiamma Buttitta, Antonio Marchetti
Pathologica.2024; 116(4): 222. CrossRef - Impact of Prolonged Ischemia on the Immunohistochemical Expression of Programmed Death Ligand 1 (PD-L1)
Angels Barberà, Juan González, Montserrat Martin, Jose L. Mate, Albert Oriol, Fina Martínez-Soler, Tomas Santalucia, Pedro Luis Fernández
Applied Immunohistochemistry & Molecular Morphology.2023; 31(9): 607. CrossRef - A practical approach for PD-L1 evaluation in gastroesophageal cancer
Valentina Angerilli, Matteo Fassan, Paola Parente, Irene Gullo, Michela Campora, Chiara Rossi, Maria Luisa Sacramento, Gianmaria Pennelli, Alessandro Vanoli, Federica Grillo, Luca Mastracci
Pathologica.2023; 115(2): 57. CrossRef - EZH2 and POU2F3 Can Aid in the Distinction of Thymic Carcinoma from Thymoma
Julia R. Naso, Julie A. Vrana, Justin W. Koepplin, Julian R. Molina, Anja C. Roden
Cancers.2023; 15(8): 2274. CrossRef - Artificial intelligence-assisted system for precision diagnosis of PD-L1 expression in non-small cell lung cancer
Jianghua Wu, Changling Liu, Xiaoqing Liu, Wei Sun, Linfeng Li, Nannan Gao, Yajun Zhang, Xin Yang, Junjie Zhang, Haiyue Wang, Xinying Liu, Xiaozheng Huang, Yanhui Zhang, Runfen Cheng, Kaiwen Chi, Luning Mao, Lixin Zhou, Dongmei Lin, Shaoping Ling
Modern Pathology.2022; 35(3): 403. CrossRef - Immunohistochemistry as predictive and prognostic markers for gastrointestinal malignancies
Matthew W. Rosenbaum, Raul S. Gonzalez
Seminars in Diagnostic Pathology.2022; 39(1): 48. CrossRef - Gastric Cancer: Mechanisms, Biomarkers, and Therapeutic Approaches
Sangjoon Choi, Sujin Park, Hyunjin Kim, So Young Kang, Soomin Ahn, Kyoung-Mee Kim
Biomedicines.2022; 10(3): 543. CrossRef - Development and validation of a supervised deep learning algorithm for automated whole‐slide programmed death‐ligand 1 tumour proportion score assessment in non‐small cell lung cancer
Liesbeth M Hondelink, Melek Hüyük, Pieter E Postmus, Vincent T H B M Smit, Sami Blom, Jan H von der Thüsen, Danielle Cohen
Histopathology.2022; 80(4): 635. CrossRef - 5-hmC loss is another useful tool in addition to BAP1 and MTAP immunostains to distinguish diffuse malignant peritoneal mesothelioma from reactive mesothelial hyperplasia in peritoneal cytology cell-blocks and biopsies
Ziyad Alsugair, Vahan Kepenekian, Tanguy Fenouil, Olivier Glehen, Laurent Villeneuve, Sylvie Isaac, Juliette Hommell-Fontaine, Nazim Benzerdjeb
Virchows Archiv.2022; 481(1): 23. CrossRef - Artificial intelligence–powered programmed death ligand 1 analyser reduces interobserver variation in tumour proportion score for non–small cell lung cancer with better prediction of immunotherapy response
Sangjoon Choi, Soo Ick Cho, Minuk Ma, Seonwook Park, Sergio Pereira, Brian Jaehong Aum, Seunghwan Shin, Kyunghyun Paeng, Donggeun Yoo, Wonkyung Jung, Chan-Young Ock, Se-Hoon Lee, Yoon-La Choi, Jin-Haeng Chung, Tony S. Mok, Hyojin Kim, Seokhwi Kim
European Journal of Cancer.2022; 170: 17. CrossRef - Artificial Intelligence-Assisted Score Analysis for Predicting the Expression of the Immunotherapy Biomarker PD-L1 in Lung Cancer
Guoping Cheng, Fuchuang Zhang, Yishi Xing, Xingyi Hu, He Zhang, Shiting Chen, Mengdao Li, Chaolong Peng, Guangtai Ding, Dadong Zhang, Peilin Chen, Qingxin Xia, Meijuan Wu
Frontiers in Immunology.2022;[Epub] CrossRef - Association of artificial intelligence-powered and manual quantification of programmed death-ligand 1 (PD-L1) expression with outcomes in patients treated with nivolumab ± ipilimumab
Vipul Baxi, George Lee, Chunzhe Duan, Dimple Pandya, Daniel N. Cohen, Robin Edwards, Han Chang, Jun Li, Hunter Elliott, Harsha Pokkalla, Benjamin Glass, Nishant Agrawal, Abhik Lahiri, Dayong Wang, Aditya Khosla, Ilan Wapinski, Andrew Beck, Michael Montalt
Modern Pathology.2022; 35(11): 1529. CrossRef - High interobserver and intraobserver reproducibility among pathologists assessing PD‐L1 CPS across multiple indications
Shanthy Nuti, Yiwei Zhang, Nabila Zerrouki, Charlotte Roach, Gudrun Bänfer, George L Kumar, Edward Manna, Rolf Diezko, Kristopher Kersch, Josef Rüschoff, Bharat Jasani
Histopathology.2022; 81(6): 732. CrossRef - Modifying factors of PD‐L1 expression on tumor cells in advanced non‐small‐cell lung cancer
Alejandro Avilés‐Salas, Diana Flores‐Estrada, Luis Lara‐Mejía, Rodrigo Catalán, Graciela Cruz‐Rico, Mario Orozco‐Morales, David Heredia, Laura Bolaño‐Guerra, Pamela Denisse Soberanis‐Piña, Edgar Varela‐Santoyo, Andrés F. Cardona, Oscar Arrieta
Thoracic Cancer.2022; 13(23): 3362. CrossRef - Comparability of laboratory-developed and commercial PD-L1 assays in non-small cell lung carcinoma
Julia R. Naso, Gang Wang, Norbert Banyi, Fatemeh Derakhshan, Aria Shokoohi, Cheryl Ho, Chen Zhou, Diana N. Ionescu
Annals of Diagnostic Pathology.2021; 50: 151590. CrossRef - Interobserver agreement in programmed cell death‐ligand 1 immunohistochemistry scoring in nonsmall cell lung carcinoma cytologic specimens
William Sinclair, Peter Kobalka, Rongqin Ren, Boulos Beshai, Abberly A. Lott Limbach, Lai Wei, Ping Mei, Zaibo Li
Diagnostic Cytopathology.2021; 49(2): 219. CrossRef - Automated PD-L1 Scoring for Non-Small Cell Lung Carcinoma Using Open-Source Software
Julia R. Naso, Tetiana Povshedna, Gang Wang, Norbert Banyi, Calum MacAulay, Diana N. Ionescu, Chen Zhou
Pathology and Oncology Research.2021;[Epub] CrossRef - The Immunohistochemical Expression of Programmed Death Ligand 1 (PD-L1) Is Affected by Sample Overfixation
Angels Barberà, Ruth Marginet Flinch, Montserrat Martin, Jose L. Mate, Albert Oriol, Fina Martínez-Soler, Tomas Santalucia, Pedro L. Fernández
Applied Immunohistochemistry & Molecular Morphology.2021; 29(1): 76. CrossRef - Programmed cell death-ligand 1 assessment in urothelial carcinoma: prospect and limitation
Kyu Sang Lee, Gheeyoung Choe
Journal of Pathology and Translational Medicine.2021; 55(3): 163. CrossRef - Comparison of Semi-Quantitative Scoring and Artificial Intelligence Aided Digital Image Analysis of Chromogenic Immunohistochemistry
János Bencze, Máté Szarka, Balázs Kóti, Woosung Seo, Tibor G. Hortobágyi, Viktor Bencs, László V. Módis, Tibor Hortobágyi
Biomolecules.2021; 12(1): 19. CrossRef - Immunization against ROS1 by DNA Electroporation Impairs K-Ras-Driven Lung Adenocarcinomas
Federica Riccardo, Giuseppina Barutello, Angela Petito, Lidia Tarone, Laura Conti, Maddalena Arigoni, Chiara Musiu, Stefania Izzo, Marco Volante, Dario Livio Longo, Irene Fiore Merighi, Mauro Papotti, Federica Cavallo, Elena Quaglino
Vaccines.2020; 8(2): 166. CrossRef - Utility of PD-L1 testing on non-small cell lung cancer cytology specimens: An institutional experience with interobserver variability analysis
Oleksandr Kravtsov, Christopher P. Hartley, Yuri Sheinin, Bryan C. Hunt, Juan C. Felix, Tamara Giorgadze
Annals of Diagnostic Pathology.2020; 48: 151602. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure


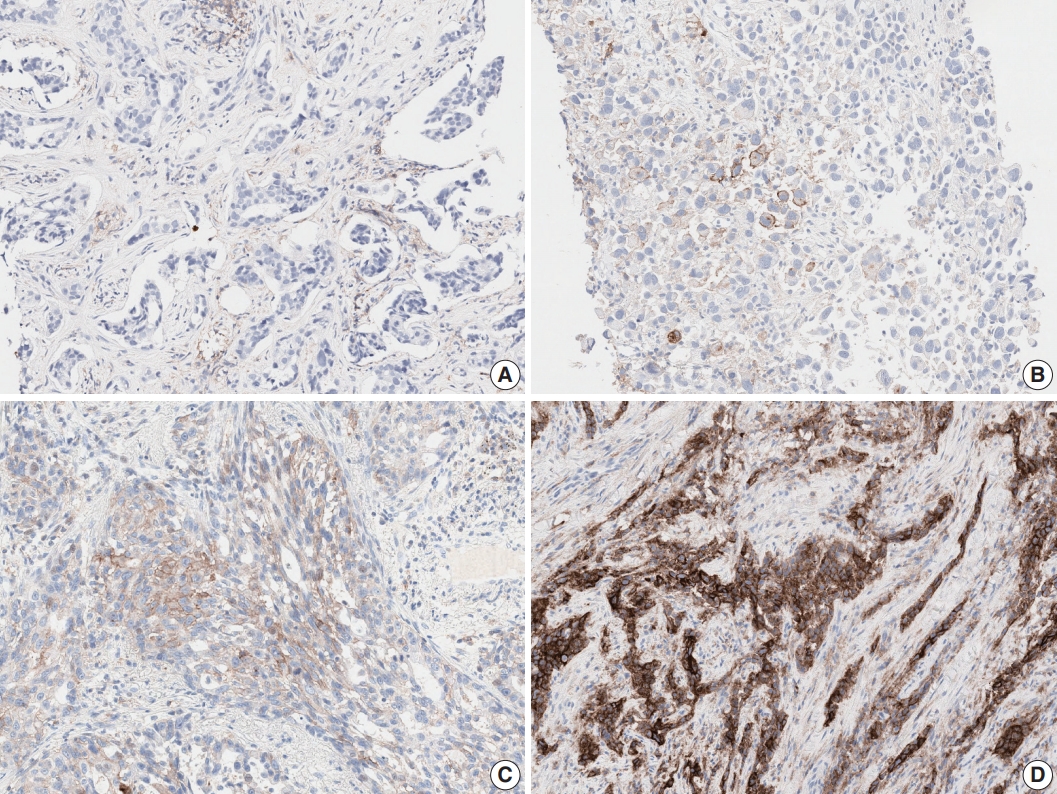
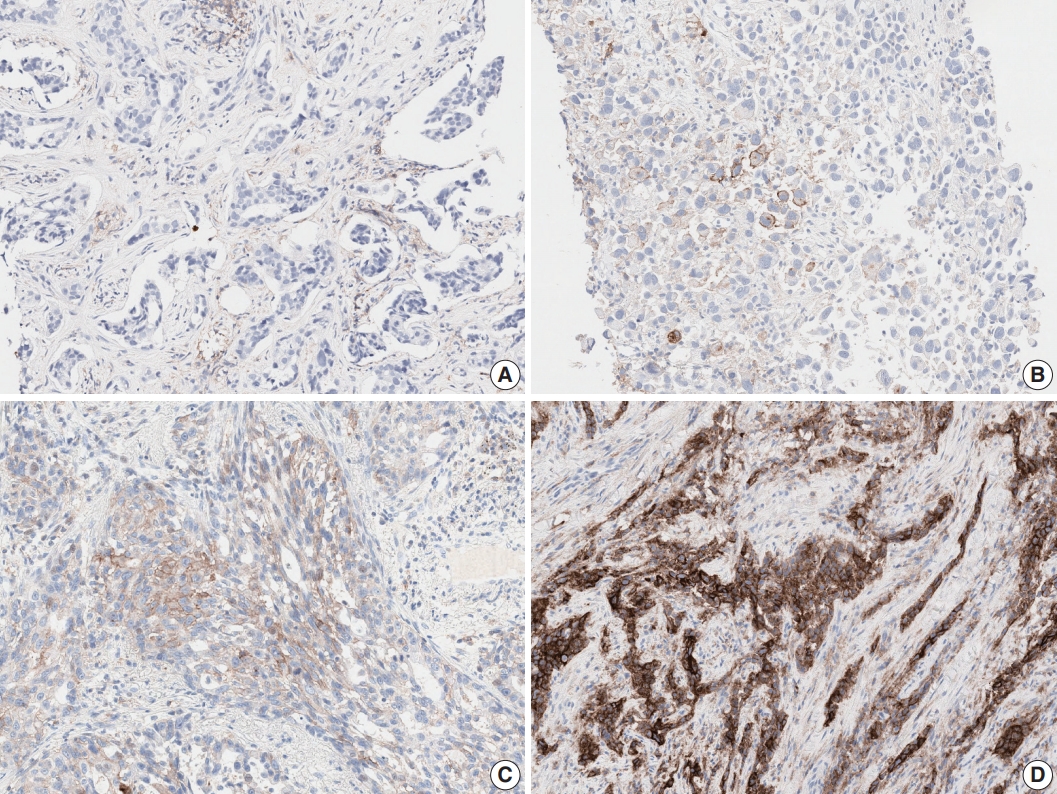
Fig. 1.
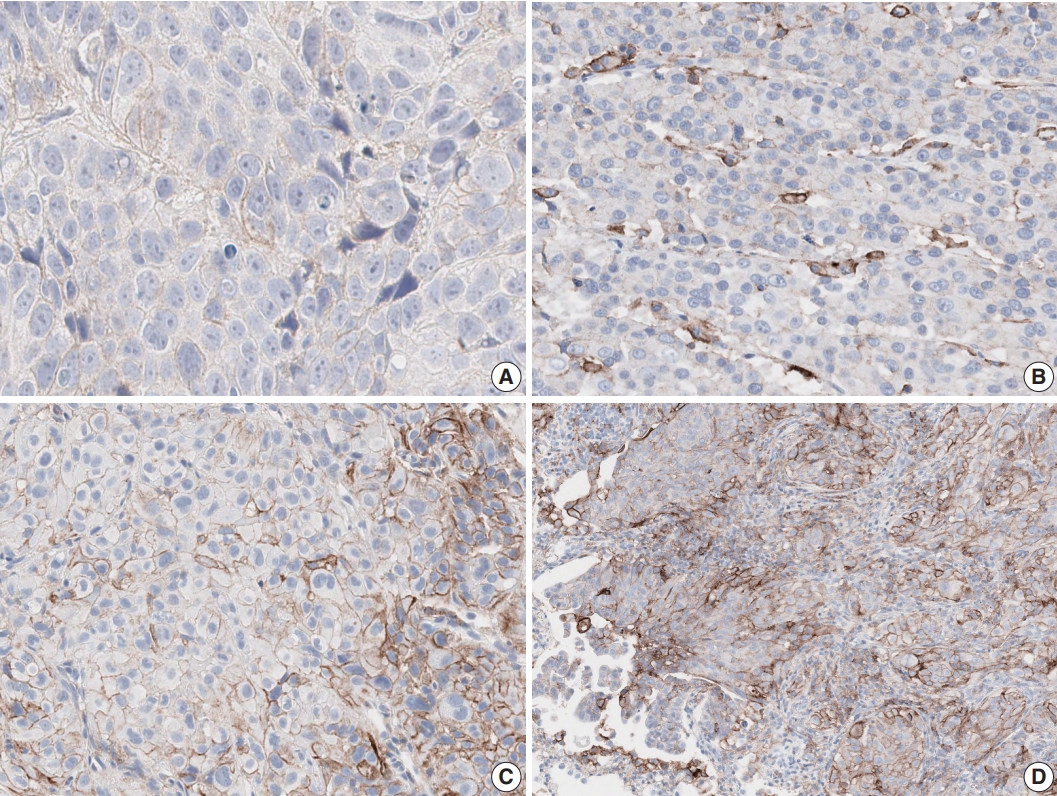
Fig. 2.
| ICC for TPS | p-value | Weighted Kappa for 3 tiered assessment | p-value | |
|---|---|---|---|---|
| Total (n=26) | 0.902 ± 0.058 | .037 | 0.748 ± 0.093 | .043 |
| Expert (n=19) | 0.919 ± 0.034 | 0.772 ± 0.073 | ||
| Fellow (n=7) | 0.859 ± 0.088 | 0.680 ± 0.115 |
| 1% Cut-off | p-value | 50% Cut-off | p-value | |
|---|---|---|---|---|
| Cohen’s κ coefficient | ||||
| Total (n = 26) | 0.633 ± 0.111 | .068 | 0.834 ± 0.095 | .082 |
| Expert (n = 19) | 0.656 ± 0.104 | 0.858 ± 0.072 | ||
| Fellow (n = 7) | 0.570 ± 0.113 | 0.768 ± 0.123 | ||
| OPA (%) | ||||
| Total (n = 26) | 86.2 ± 5.5 | .067 | 92.1 ± 4.4 | .075 |
| Expert (n = 19) | 87.3 ± 4.6 | 93.2 ± 3.3 | ||
| Fellow (n = 7) | 83.2 ± 6.8 | 89.1 ± 5.5 | ||
| NPA (%) | ||||
| Total (n = 26) | 85.7 ± 16.0 | .150 | 95.4 ± 4.3 | .884 |
| Expert (n = 19) | 86.8 ± 17.1 | 95.3 ± 4.5 | ||
| Fellow (n = 7) | 82.5 ± 13.5 | 95.4 ± 4.0 | ||
| PPA (%) | ||||
| Total (n = 26) | 86.3 ± 8.4 | .385 | 87.5 ± 12.0 | .149 |
| Expert (n = 19) | 87.4 ± 7.6 | 9.2 ± 9.2 | ||
| Fellow (n = 7) | 83.4 ± 1.2 | 8.3 ± 16.4 |
| EBUS-TBNA (n = 19) | Biopsy (n = 66) | Resection (n = 22) | p-value | |
|---|---|---|---|---|
| ICC for TPS | 0.887 ± 0.099 | 0.899 ± 0.061 | 0.926 ± 0.050 | .023 |
| κ for 3-step evaluation | 0.669 ± 0.089 | 0.776 ± 0.104 | 0.716 ± 0.126 | .001 |
| κ for 1% cutoff | 0.383 ± 0.134 | 0.713 ± 0.137 | 0.538 ± 0.209 | |
| κ for 50% cutoff | 0.832 ± 0.128 | 0.830 ± 0.097 | 0.839 ± 0.118 |
| SCC (n = 33) | ADC (n = 66) | p-value | |
|---|---|---|---|
| ICC for TPS | 0.877 ± 0.071 | 0.917 ± 0.053 | .024 |
| κ for 3-step evaluation | 0.757 ± 0.107 | 0.753 ± 0.089 | .073 |
| Training |
p-value | ||
|---|---|---|---|
| Yes (n = 15) | No (n = 11) | ||
| ICC for TPS | 0.922 ± 0.034 | 0.875 ± 0.074 | .043 |
| κ for 3-step assessment | 0.776 ± 0.079 | 0.709 ± 0.101 | .026 |
| ICC for TPS |
κ coefficient for 3-step evaluation |
|||
|---|---|---|---|---|
| Spearman correlation | p-value | Spearman correlation | p-value | |
| PD-L1 test experience | 0.422 | .032 | 0.277 | .170 |
| Practice duration | 0.477 | .014 | 0.527 | .005 |
| Sample | Observer | 1% Cut-off | 50% Cut-off | |
|---|---|---|---|---|
| Rimm et al. [8] | 90 | 13 | κ = 0.537 |
κ = 0.749 |
| Cooper et al. [9] | 108 | 10 | κ = 0.68 | κ = 0.58 |
| Brunnström et al. [10] | 55 | 7 | 2%–20% |
0%–2% |
| Current study | 107 | 27 | κ = 0.633 | κ = 0.834 |
ICC, Intraclass correlation coefficient; TPS, tumor proportion score.
OPA, overall percent agreement; NPA, negative percent agreement; PPA, positive percent agreement.
EBUS-TBNA, Endobronchial ultrasound-guided transbronchial needle aspiration; ICC, intraclass correlation coefficient; TPS, tumor proportion score.
SCC, squamous cell carcinoma; ADC, adenocarcinoma; ICC, intraclass correlation coefficient; TPS, tumor proportion score.
ICC, intraclass correlation coefficient; TPS, tumor proportion score.
ICC, intraclass correlation coefficient; TPS, tumor proportion score; PD-L1, programmed cell death-ligand 1.
For the mean of all four assay (22C3, 28-8, SP142, and E1L3N); Differently classified cases by any one pathologist compared with consensus.

 E-submission
E-submission