Search
- Page Path
- HOME > Search
- Clinical practice recommendations for the use of next-generation sequencing in patients with solid cancer: a joint report from KSMO and KSP
- Miso Kim, Hyo Sup Shim, Sheehyun Kim, In Hee Lee, Jihun Kim, Shinkyo Yoon, Hyung-Don Kim, Inkeun Park, Jae Ho Jeong, Changhoon Yoo, Jaekyung Cheon, In-Ho Kim, Jieun Lee, Sook Hee Hong, Sehhoon Park, Hyun Ae Jung, Jin Won Kim, Han Jo Kim, Yongjun Cha, Sun Min Lim, Han Sang Kim, Choong-Kun Lee, Jee Hung Kim, Sang Hoon Chun, Jina Yun, So Yeon Park, Hye Seung Lee, Yong Mee Cho, Soo Jeong Nam, Kiyong Na, Sun Och Yoon, Ahwon Lee, Kee-Taek Jang, Hongseok Yun, Sungyoung Lee, Jee Hyun Kim, Wan-Seop Kim
- J Pathol Transl Med. 2024;58(4):147-164. Published online January 10, 2024
- DOI: https://doi.org/10.4132/jptm.2023.11.01
- 8,554 View
- 494 Download
- 1 Web of Science
- 2 Crossref
-
 Abstract
Abstract
 PDF
PDF - In recent years, next-generation sequencing (NGS)–based genetic testing has become crucial in cancer care. While its primary objective is to identify actionable genetic alterations to guide treatment decisions, its scope has broadened to encompass aiding in pathological diagnosis and exploring resistance mechanisms. With the ongoing expansion in NGS application and reliance, a compelling necessity arises for expert consensus on its application in solid cancers. To address this demand, the forthcoming recommendations not only provide pragmatic guidance for the clinical use of NGS but also systematically classify actionable genes based on specific cancer types. Additionally, these recommendations will incorporate expert perspectives on crucial biomarkers, ensuring informed decisions regarding circulating tumor DNA panel testing.
-
Citations
Citations to this article as recorded by- Apport de la génomique dans la prise en charge des cancers
Étienne Rouleau, Lucie Karayan-Tapon, Marie-Dominique Galibert, Alexandre Harlé, Isabelle Soubeyran
Revue Francophone des Laboratoires.2025; 2025(568): 67. CrossRef - The Redox–Adhesion–Exosome (RAX) Hub in Cancer: Lipid Peroxidation-Driven EMT Plasticity and Ferroptosis Defense with HNE/MDA Signaling and Lipidomic Perspectives
Moon Nyeo Park, Jinwon Choi, Rosy Iara Maciel de Azambuja Ribeiro, Domenico V. Delfino, Seong-Gyu Ko, Bonglee Kim
Antioxidants.2025; 14(12): 1474. CrossRef
- Apport de la génomique dans la prise en charge des cancers
- Establishing molecular pathology curriculum for pathology trainees and continued medical education: a collaborative work from the Molecular Pathology Study Group of the Korean Society of Pathologists
- Jiwon Koh, Ha Young Park, Jeong Mo Bae, Jun Kang, Uiju Cho, Seung Eun Lee, Haeyoun Kang, Min Eui Hong, Jae Kyung Won, Youn-La Choi, Wan-Seop Kim, Ahwon Lee
- J Pathol Transl Med. 2023;57(5):265-272. Published online September 15, 2023
- DOI: https://doi.org/10.4132/jptm.2023.08.26
- 5,399 View
- 207 Download
- 1 Web of Science
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF - Background
The importance of molecular pathology tests has increased during the last decade, and there is a great need for efficient training of molecular pathology for pathology trainees and as continued medical education.
Methods
The Molecular Pathology Study Group of the Korean Society of Pathologists appointed a task force composed of experienced molecular pathologists to develop a refined educational curriculum of molecular pathology. A 3-day online educational session was held based on the newly established structure of learning objectives; the audience were asked to score their understanding of 22 selected learning objectives before and after the session to assess the effect of structured education.
Results
The structured objectives and goals of molecular pathology was established and posted as a web-based interface which can serve as a knowledge bank of molecular pathology. A total of 201 pathologists participated in the educational session. For all 22 learning objectives, the scores of self-reported understanding increased after educational session by 9.9 points on average (range, 6.6 to 17.0). The most effectively improved items were objectives from next-generation sequencing (NGS) section: ‘NGS library preparation and quality control’ (score increased from 51.8 to 68.8), ‘NGS interpretation of variants and reference database’ (score increased from 54.1 to 68.0), and ‘whole genome, whole exome, and targeted gene sequencing’ (score increased from 58.2 to 71.2). Qualitative responses regarding the adequacy of refined educational curriculum were collected, where favorable comments dominated.
Conclusions
Approach toward the education of molecular pathology was refined, which would greatly benefit the future trainees. -
Citations
Citations to this article as recorded by- Presence of RB1 or Absence of LRP1B Mutation Predicts Poor Overall Survival in Patients with Gastric Neuroendocrine Carcinoma and Mixed Adenoneuroendocrine Carcinoma
In Hye Song, Bokyung Ahn, Young Soo Park, Deok Hoon Kim, Seung-Mo Hong
Cancer Research and Treatment.2025; 57(2): 492. CrossRef
- Presence of RB1 or Absence of LRP1B Mutation Predicts Poor Overall Survival in Patients with Gastric Neuroendocrine Carcinoma and Mixed Adenoneuroendocrine Carcinoma
- Single-center study on clinicopathological and typical molecular pathologic features of metastatic brain tumor
- Su Hwa Kim, Young Suk Lee, Sung Hak Lee, Yeoun Eun Sung, Ahwon Lee, Jun Kang, Jae-Sung Park, Sin Soo Jeun, Youn Soo Lee
- J Pathol Transl Med. 2023;57(4):217-231. Published online July 11, 2023
- DOI: https://doi.org/10.4132/jptm.2023.06.10
- 5,368 View
- 173 Download
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF - Background
The metastatic brain tumor is the most common brain tumor. The aim of this study was to demonstrate the clinicopathological and molecular pathologic features of brain metastases (BM).
Methods
A total of 269 patients were diagnosed with BM through surgical resection at Seoul St. Mary’s Hospital from January 2010 to March 2020. We reviewed the clinicopathological features and molecular status of primary and metastatic brain tissues using immunohistochemistry and molecular pathology results.
Results
Among 269 patients, 139 males and 130 females were included. The median age of primary tumor was 58 years (range, 13 to 87 years) and 86 patients (32.0%) had BM at initial presentation. Median BM free interval was 28.0 months (range, 1 to 286 months). The most frequent primary site was lung 46.5% (125/269), and followed by breast 15.6% (42/269), colorectum 10.0% (27/269). Epidermal growth factor receptor (EGFR) mutation was found in 50.8% (32/63) and 58.0% (40/69) of lung primary and BM, respectively. In both breast primary and breast cancer with BM, luminal B was the most frequent subtype at 37.9% (11/29) and 42.9% (18/42), respectively, followed by human epidermal growth factor receptor 2 with 31.0% (9/29) and 33.3% (14/42). Triple-negative was 20.7% (6/29) and 16.7% (7/42), and luminal A was 10.3% (3/29) and 7.1% (3/42) of breast primary and BM, respectively. In colorectal primary and colorectal cancer with BM, KRAS mutation was found in 76.9% (10/13) and 66.7% (2/3), respectively.
Conclusions
We report the clinicopathological and molecular pathologic features of BM that can provide useful information for understanding the pathogenesis of metastasis and for clinical trials based on the tumor’s molecular pathology. -
Citations
Citations to this article as recorded by- Colorectal cancer metastasis to the brain: A scoping review of incidence, treatment, and outcomes
Hunter J Hutchinson, Melanie Gonzalez, Diana Feier, Colin E Welch, Brandon Lucke-Wold
World Journal of Gastrointestinal Pathophysiology.2025;[Epub] CrossRef
- Colorectal cancer metastasis to the brain: A scoping review of incidence, treatment, and outcomes
- A multicenter study of interobserver variability in pathologic diagnosis of papillary breast lesions on core needle biopsy with WHO classification
- Hye Ju Kang, Sun Young Kwon, Ahrong Kim, Woo Gyeong Kim, Eun Kyung Kim, Ae Ree Kim, Chungyeul Kim, Soo Kee Min, So Young Park, Sun Hee Sung, Hye Kyoung Yoon, Ahwon Lee, Ji Shin Lee, Hyang Im Lee, Ho Chang Lee, Sung Chul Lim, Sun Young Jun, Min Jung Jung, Chang Won Jung, Soo Youn Cho, Eun Yoon Cho, Hye Jeong Choi, So Yeon Park, Jee Yeon Kim, In Ae Park, Youngmee Kwon
- J Pathol Transl Med. 2021;55(6):380-387. Published online October 6, 2021
- DOI: https://doi.org/10.4132/jptm.2021.07.29
- 7,212 View
- 226 Download
- 4 Web of Science
- 5 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
Papillary breast lesions (PBLs) comprise diverse entities from benign and atypical lesions to malignant tumors. Although PBLs are characterized by a papillary growth pattern, it is challenging to achieve high diagnostic accuracy and reproducibility. Thus, we investigated the diagnostic reproducibility of PBLs in core needle biopsy (CNB) specimens with World Health Organization (WHO) classification.
Methods
Diagnostic reproducibility was assessed using interobserver variability (kappa value, κ) and agreement rate in the pathologic diagnosis of 60 PBL cases on CNB among 20 breast pathologists affiliated with 20 medical institutions in Korea. This analysis was performed using hematoxylin and eosin (H&E) staining and immunohistochemical (IHC) staining for cytokeratin 5 (CK5) and p63. The pathologic diagnosis of PBLs was based on WHO classification, which was used to establish simple classifications (4-tier, 3-tier, and 2-tier).
Results
On WHO classification, H&E staining exhibited ‘fair agreement’ (κ = 0.21) with a 47.0% agreement rate. Simple classifications presented improvement in interobserver variability and agreement rate. IHC staining increased the kappa value and agreement rate in all the classifications. Despite IHC staining, the encapsulated/solid papillary carcinoma (EPC/SPC) subgroup (κ = 0.16) exhibited lower agreement compared to the non-EPC/SPC subgroup (κ = 0.35) with WHO classification, which was similar to the results of any other classification systems.
Conclusions
Although the use of IHC staining for CK5 and p63 increased the diagnostic agreement of PBLs in CNB specimens, WHO classification exhibited a higher discordance rate compared to any other classifications. Therefore, this result warrants further intensive consensus studies to improve the diagnostic reproducibility of PBLs with WHO classification. -
Citations
Citations to this article as recorded by- Beyond the benign: A rare case report of myxoid pleomorphic liposarcoma
Arslan Ahmad, Muhammad Ammar, Muhammad Hasnain Saleem Choudary, Muhammad Nouman Sadiq, Rana Uzair Ahmad, Nouman Aziz
Radiology Case Reports.2025; 20(5): 2500. CrossRef - Invasive papillary carcinoma of the breast
Shijing Wang, Qingfu Zhang, Xiaoyun Mao
Frontiers in Oncology.2024;[Epub] CrossRef - Recommendations for Performance Evaluation of Machine Learning in Pathology: A Concept Paper From the College of American Pathologists
Matthew G. Hanna, Niels H. Olson, Mark Zarella, Rajesh C. Dash, Markus D. Herrmann, Larissa V. Furtado, Michelle N. Stram, Patricia M. Raciti, Lewis Hassell, Alex Mays, Liron Pantanowitz, Joseph S. Sirintrapun, Savitri Krishnamurthy, Anil Parwani, Giovann
Archives of Pathology & Laboratory Medicine.2024; 148(10): e335. CrossRef - Encapsulated papillary carcinoma of the breast: A single institution experience
Liang Xu, Qixin Mao, Qiuming Liu, Yufeng Gao, Lihua Luo, Chungen Guo, Wei Qu, Ningning Yan, Yali Cao
Oncology Letters.2023;[Epub] CrossRef - High-risk and selected benign breast lesions diagnosed on core needle biopsy: Evidence for and against immediate surgical excision
Aparna Harbhajanka, Hannah L. Gilmore, Benjamin C. Calhoun
Modern Pathology.2022; 35(11): 1500. CrossRef
- Beyond the benign: A rare case report of myxoid pleomorphic liposarcoma
- Standardized pathology report for breast cancer
- Soo Youn Cho, So Yeon Park, Young Kyung Bae, Jee Yeon Kim, Eun Kyung Kim, Woo Gyeong Kim, Youngmee Kwon, Ahwon Lee, Hee Jin Lee, Ji Shin Lee, Jee Young Park, Gyungyub Gong, Hye Kyoung Yoon
- J Pathol Transl Med. 2021;55(1):1-15. Published online January 11, 2021
- DOI: https://doi.org/10.4132/jptm.2020.11.20
- 15,907 View
- 725 Download
- 8 Web of Science
- 4 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Given the recent advances in management and understanding of breast cancer, a standardized pathology report reflecting these changes is critical. To meet this need, the Breast Pathology Study Group of the Korean Society of Pathologists has developed a standardized pathology reporting format for breast cancer, consisting of ‘standard data elements,’ ‘conditional data elements,’ and a biomarker report form. The ‘standard data elements’ consist of the basic pathologic features used for prognostication, while other factors related to prognosis or diagnosis are described in the ‘conditional data elements.’ In addition to standard data elements, all recommended issues are also presented. We expect that this standardized pathology report for breast cancer will improve diagnostic concordance and communication between pathologists and clinicians, as well as between pathologists inter-institutionally.
-
Citations
Citations to this article as recorded by- Adenoid Cystic Carcinoma of Breast Associated With an Incidental Radial Scar: A Cyto‐Histopathology Correlation
Rallapalli Rajyalakshmi, Valasapalli Rajani, Tanuku Sreedhar, Kollabathula Arpitha
Diagnostic Cytopathology.2026;[Epub] CrossRef - Navigating discrepancies: The assessment of residual lymphovascular invasion in breast carcinoma after neoadjuvant treatment
Anikó Kovács, Åsa Rundgren-Sellei, Gunilla Rask, Annette Bauer, Anna Bodén, Johannes van Brakel, Eugenia Colón-Cervantes, Anna Ehinger, Johan Hartman, Balazs Acs
The Breast.2025; 82: 104519. CrossRef - Residual pure intralymphatic carcinoma component only (lymphovascular tumor emboli without invasive carcinoma) after neoadjuvant chemotherapy is associated with poor outcome: Not pathologic complete response
Hyunwoo Lee, Yunjeong Jang, Yoon Ah Cho, Eun Yoon Cho
Human Pathology.2024; 145: 1. CrossRef - Sentinel lymph node biopsy in patients with ductal carcinomain situ: systematic review and meta-analysis
Matthew G. Davey, Colm O’Flaherty, Eoin F. Cleere, Aoife Nohilly, James Phelan, Evan Ronane, Aoife J. Lowery, Michael J. Kerin
BJS Open.2022;[Epub] CrossRef
- Adenoid Cystic Carcinoma of Breast Associated With an Incidental Radial Scar: A Cyto‐Histopathology Correlation
- Analysis of the molecular subtypes of preoperative core needle biopsy and surgical specimens in invasive breast cancer
- Ye Sul Jeong, Jun Kang, Jieun Lee, Tae-Kyung Yoo, Sung Hun Kim, Ahwon Lee
- J Pathol Transl Med. 2020;54(1):87-94. Published online November 13, 2019
- DOI: https://doi.org/10.4132/jptm.2019.10.14
- 10,152 View
- 212 Download
- 18 Web of Science
- 20 Crossref
-
 Abstract
Abstract
 PDF
PDF - Background
Accurate molecular classification of breast core needle biopsy (CNB) tissue is important for determining neoadjuvant systemic therapies for invasive breast cancer. The researchers aimed to evaluate the concordance rate (CR) of molecular subtypes between CNBs and surgical specimens.
Methods
This study was conducted with invasive breast cancer patients who underwent surgery after CNB at Seoul St. Mary’s Hospital between December 2014 and December 2017. Estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and Ki67 were analyzed using immunohistochemistry. ER and PR were evaluated by Allred score (0–8). HER2 was graded from 0 to +3, and all 2+ cases were reflex tested with silver in situ hybridization. The labeling index of Ki67 was counted by either manual scoring or digital image analysis. Molecular subtypes were classified using the above surrogate markers.
Results
In total, 629 patients were evaluated. The CRs of ER, PR, HER2, and Ki67 were 96.5% (kappa, 0.883; p<.001), 93.0% (kappa, 0.824; p<.001), 99.7% (kappa, 0.988; p<.001), and 78.7% (kappa, 0.577; p<.001), respectively. Digital image analysis of Ki67 in CNB showed better concordance with Ki67 in surgical specimens (CR, 82.3%; kappa, 0.639 for digital image analysis vs. CR, 76.2%; kappa, 0.534 for manual counting). The CRs of luminal A, luminal B, HER2, and triple negative types were 89.0%, 70.0%, 82.9%, and 77.2%, respectively.
Conclusions
CNB was reasonably accurate for determining ER, PR, HER2, Ki67, and molecular subtypes. Using digital image analysis for Ki67 in CNB produced more accurate molecular classifications. -
Citations
Citations to this article as recorded by- Predicting the Efficacy of Breast Cancer Neoadjuvant Chemotherapy Using Ultrasonography and Machine Learning
Meihong Jia, Huizhan Li, Wenli Xiao, Jiping Xue, Zhifen Wang, Xia He, Xin Wang, Dianxia Men
Ultrasound in Medicine & Biology.2026;[Epub] CrossRef - Correlation between ultrasonography and elastography parameters and molecular subtypes of breast cancer in young women
Dian-xia Men, Hui-zhan Li, Juan Dong, Meng-hua Xue, Zhi-fen Wang, Wen-li Xiao, Ji-ping Xue, Mei-hong Jia
Annals of Medicine.2025;[Epub] CrossRef - Concordance of Oncotype DX Breast Recurrence Score Assay Results Between Paired Core Needle Biopsy and Surgical Excision Specimens in Hormone Receptor Positive, Human Epidermal Growth Factor Receptor 2 Negative, Early-Stage Breast Cancer
Aziza Nassar, Jodi Carter, Paige Innis, Andrea Pingitore Blacklock, Jennifer Racz, Matthew Petitt, Purva Singla, Helena Hanna, Abigail Lochala, Christy A. Russell, Minetta C. Liu
JCO Precision Oncology.2025;[Epub] CrossRef - Impact of immunohistochemistry staining conditions on the incidence of human epidermal growth factor receptor 2 (HER2)-low breast cancer
Min Chong Kim, Sun Young Kwon, Hye Ra Jung, Young Kyung Bae
Virchows Archiv.2024; 485(6): 1117. CrossRef - Study on Intratumoral Heterogeneity of Expression of Estrogen Receptor, Progesterone Receptor, and Human Epidermal Growth Factor Receptor 2 in Carcinoma Breast
Ragavi Uthayasuriyan, Sheba K Jacob, Saloni Naresh Shah
Apollo Medicine.2024; 21(1): 51. CrossRef - Concordance of HER2 status between core needle biopsy and surgical resection specimens of breast cancer: an analysis focusing on the HER2-low status
Sei Na, Milim Kim, Yujun Park, Hyun Jung Kwon, Hee-Chul Shin, Eun-Kyu Kim, Mijung Jang, Sun Mi Kim, So Yeon Park
Breast Cancer.2024; 31(4): 705. CrossRef - Concordance of immunohistochemistry for predictive and prognostic factors in breast cancer between biopsy and surgical excision: a single-centre experience and review of the literature
Chiara Rossi, Sara Fraticelli, Marianna Fanizza, Alberta Ferrari, Elisa Ferraris, Alessia Messina, Angelica Della Valle, Chiara Annunziata Pasqualina Anghelone, Angioletta Lasagna, Gianpiero Rizzo, Lorenzo Perrone, Maria Grazia Sommaruga, Giulia Meloni, S
Breast Cancer Research and Treatment.2023; 198(3): 573. CrossRef - Single-center study on clinicopathological and typical molecular pathologic features of metastatic brain tumor
Su Hwa Kim, Young Suk Lee, Sung Hak Lee, Yeoun Eun Sung, Ahwon Lee, Jun Kang, Jae-Sung Park, Sin Soo Jeun, Youn Soo Lee
Journal of Pathology and Translational Medicine.2023; 57(4): 217. CrossRef - The Role of Diffusion-Weighted Imaging Based on Maximum-Intensity Projection in Young Patients with Marked Background Parenchymal Enhancement on Contrast-Enhanced Breast MRI
Ga-Eun Park, Bong-Joo Kang, Sung-hun Kim, Na-Young Jung
Life.2023; 13(8): 1744. CrossRef - Concordance between core needle biopsy and surgical excision specimens for Ki‐67 in breast cancer – a systematic review of the literature
Jahnavi Kalvala, Ruth M Parks, Andrew R Green, Kwok‐Leung Cheung
Histopathology.2022; 80(3): 468. CrossRef - İnvaziv Meme Kanserinde Preoperatif Kor İğne Biyopsi ile Postoperatif Cerrahi Spesmenler Arasında ER, PR, HER2 ve Ki67 Açısından Karşılaştırma
Pınar CELEPLİ, Pelin Seher ÖZTEKİN, Salih CELEPLİ, İrem BİGAT, Sema HÜCÜMENOĞLU
Akdeniz Medical Journal.2022; : 179. CrossRef - Concordance of breast cancer biomarker testing in core needle biopsy and surgical specimens: A single institution experience
Jessica A. Slostad, Nicole K. Yun, Aimee E. Schad, Surbhi Warrior, Louis F. Fogg, Ruta Rao
Cancer Medicine.2022; 11(24): 4954. CrossRef - N-Cadherin Distinguishes Intrahepatic Cholangiocarcinoma from Liver Metastases of Ductal Adenocarcinoma of the Pancreas
Tiemo S. Gerber, Benjamin Goeppert, Anne Hausen, Hagen R. Witzel, Fabian Bartsch, Mario Schindeldecker, Lisa-Katharina Gröger, Dirk A. Ridder, Oscar Cahyadi, Irene Esposito, Matthias M. Gaida, Peter Schirmacher, Peter R. Galle, Hauke Lang, Wilfried Roth,
Cancers.2022; 14(13): 3091. CrossRef - Association of Ki-67 Change Pattern After Core Needle Biopsy and Prognosis in HR+/HER2− Early Breast Cancer Patients
Shuai Li, Xiaosong Chen, Kunwei Shen
Frontiers in Surgery.2022;[Epub] CrossRef - MRI Features for Prediction Malignant Intra-Mammary Lymph Nodes: Correlations with Mammography and Ultrasound
Meejung Kim, Bong Joo Kang, Ga Eun Park
Investigative Magnetic Resonance Imaging.2022; 26(2): 135. CrossRef - A single centre experience in Turkey for comparison between core needle biopsy and surgical specimen evaluation results for HER2, SISH, estrogen receptors and progesterone receptors in breast cancer patients
Hatice Karaman, Fatma Senel, Arzu Tasdemir, Ipek Özer, Merve Dogan
Journal of Cancer Research and Therapeutics.2022; 18(6): 1789. CrossRef - Meme kanseri trucut ve rezeksiyon materyallerinde yeni moleküler sınıflama, tanı ve hormon reseptörlerinin durumu tutarlı mı?
Yeliz ARMAN KARAKAYA, Sevda YILMAZ, Hande KARABAŞ
Pamukkale Medical Journal.2021;[Epub] CrossRef - What shear wave elastography parameter best differentiates breast cancer and predicts its histologic aggressiveness?
Hyunjin Kim, Jeongmin Lee, Bong Joo Kang, Sung Hun Kim
Ultrasonography.2021; 40(2): 265. CrossRef - Risk-based decision-making in the treatment of HER2-positive early breast cancer: Recommendations based on the current state of knowledge
Christian Jackisch, Patricia Cortazar, Charles E. Geyer, Luca Gianni, Joseph Gligorov, Zuzana Machackova, Edith A. Perez, Andreas Schneeweiss, Sara M. Tolaney, Michael Untch, Andrew Wardley, Martine Piccart
Cancer Treatment Reviews.2021; 99: 102229. CrossRef - Factors influencing agreement of breast cancer luminal molecular subtype by Ki67 labeling index between core needle biopsy and surgical resection specimens
Kristina A. Tendl-Schulz, Fabian Rössler, Philipp Wimmer, Ulrike M. Heber, Martina Mittlböck, Nicolas Kozakowski, Katja Pinker, Rupert Bartsch, Peter Dubsky, Florian Fitzal, Martin Filipits, Fanny Carolina Eckel, Eva-Maria Langthaler, Günther Steger, Mich
Virchows Archiv.2020; 477(4): 545. CrossRef
- Predicting the Efficacy of Breast Cancer Neoadjuvant Chemotherapy Using Ultrasonography and Machine Learning
- KRAS Mutation Test in Korean Patients with Colorectal Carcinomas: A Methodological Comparison between Sanger Sequencing and a Real-Time PCR-Based Assay
- Sung Hak Lee, Arthur Minwoo Chung, Ahwon Lee, Woo Jin Oh, Yeong Jin Choi, Youn-Soo Lee, Eun Sun Jung
- J Pathol Transl Med. 2017;51(1):24-31. Published online December 25, 2016
- DOI: https://doi.org/10.4132/jptm.2016.10.03
- 12,401 View
- 170 Download
- 5 Web of Science
- 5 Crossref
-
 Abstract
Abstract
 PDF
PDF Supplementary Material
Supplementary Material - Background
Mutations in the KRAS gene have been identified in approximately 50% of colorectal cancers (CRCs). KRAS mutations are well established biomarkers in anti–epidermal growth factor receptor therapy. Therefore, assessment of KRAS mutations is needed in CRC patients to ensure appropriate treatment.
Methods
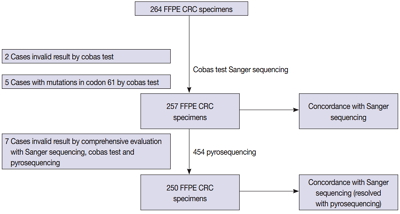
We compared the analytical performance of the cobas test to Sanger sequencing in 264 CRC cases. In addition, discordant specimens were evaluated by 454 pyrosequencing.
Results
KRAS mutations for codons 12/13 were detected in 43.2% of cases (114/264) by Sanger sequencing. Of 257 evaluable specimens for comparison, KRAS mutations were detected in 112 cases (43.6%) by Sanger sequencing and 118 cases (45.9%) by the cobas test. Concordance between the cobas test and Sanger sequencing for each lot was 93.8% positive percent agreement (PPA) and 91.0% negative percent agreement (NPA) for codons 12/13. Results from the cobas test and Sanger sequencing were discordant for 20 cases (7.8%). Twenty discrepant cases were subsequently subjected to 454 pyrosequencing. After comprehensive analysis of the results from combined Sanger sequencing–454 pyrosequencing and the cobas test, PPA was 97.5% and NPA was 100%.
Conclusions
The cobas test is an accurate and sensitive test for detecting KRAS-activating mutations and has analytical power equivalent to Sanger sequencing. Prescreening using the cobas test with subsequent application of Sanger sequencing is the best strategy for routine detection of KRAS mutations in CRC. -
Citations
Citations to this article as recorded by- Single-center study on clinicopathological and typical molecular pathologic features of metastatic brain tumor
Su Hwa Kim, Young Suk Lee, Sung Hak Lee, Yeoun Eun Sung, Ahwon Lee, Jun Kang, Jae-Sung Park, Sin Soo Jeun, Youn Soo Lee
Journal of Pathology and Translational Medicine.2023; 57(4): 217. CrossRef - Assessment of KRAS and NRAS status in metastatic colorectal cancer: Experience of the National Institute of Oncology in Rabat Morocco
Chaimaa Mounjid, Hajar El Agouri, Youssef Mahdi, Abdelilah Laraqui, En-nacer Chtati, Soumaya Ech-charif, Mouna Khmou, Youssef Bakri, Amine Souadka, Basma El Khannoussi
Annals of Cancer Research and Therapy.2022; 30(2): 80. CrossRef - The current understanding on the impact of KRAS on colorectal cancer
Mingjing Meng, Keying Zhong, Ting Jiang, Zhongqiu Liu, Hiu Yee Kwan, Tao Su
Biomedicine & Pharmacotherapy.2021; 140: 111717. CrossRef - Droplet digital PCR revealed high concordance between primary tumors and lymph node metastases in multiplex screening of KRAS mutations in colorectal cancer
Barbora Vanova, Michal Kalman, Karin Jasek, Ivana Kasubova, Tatiana Burjanivova, Anna Farkasova, Peter Kruzliak, Dietrich Busselberg, Lukas Plank, Zora Lasabova
Clinical and Experimental Medicine.2019; 19(2): 219. CrossRef - CRISPR Technology for Breast Cancer: Diagnostics, Modeling, and Therapy
Rachel L. Mintz, Madeleine A. Gao, Kahmun Lo, Yeh‐Hsing Lao, Mingqiang Li, Kam W. Leong
Advanced Biosystems.2018;[Epub] CrossRef
- Single-center study on clinicopathological and typical molecular pathologic features of metastatic brain tumor
- Clinical Significance of an HPV DNA Chip Test with Emphasis on HPV-16 and/or HPV-18 Detection in Korean Gynecological Patients
- Min-Kyung Yeo, Ahwon Lee, Soo Young Hur, Jong Sup Park
- J Pathol Transl Med. 2016;50(4):294-299. Published online June 26, 2016
- DOI: https://doi.org/10.4132/jptm.2016.05.09
- 10,652 View
- 80 Download
- 3 Web of Science
- 2 Crossref
-
 Abstract
Abstract
 PDF
PDF - Background
Human papillomavirus (HPV) is a major risk factor for cervical cancer.
Methods
We evaluated the clinical significance of the HPV DNA chip genotyping assay (MyHPV chip, Mygene Co.) compared with the Hybrid Capture 2 (HC2) chemiluminescent nucleic acid hybridization kit (Digene Corp.) in 867 patients.
Results
The concordance rate between the MyHPV chip and HC2 was 79.4% (kappa coefficient, κ = 0.55). The sensitivity and specificity of both HPV tests were very similar (approximately 85% and 50%, respectively). The addition of HPV result (either MyHPV chip or HC2) to cytology improved the sensitivity (95%, each) but reduced the specificity (approximately 30%, each) compared with the HPV test or cytology alone. Based on the MyHPV chip results, the odds ratio (OR) for ≥ high-grade squamous intraepithelial lesions (HSILs) was 9.9 in the HPV-16/18 (+) group and 3.7 in the non-16/18 high-risk (HR)-HPV (+) group. Based on the HC2 results, the OR for ≥ HSILs was 5.9 in the HR-HPV (+) group. When considering only patients with cytological diagnoses of “negative for intraepithelial lesion or malignancy” and “atypical squamous cell or atypical glandular cell,” based on the MyHPV chip results, the ORs for ≥ HSILs were 6.8 and 11.7, respectively, in the HPV-16/18 (+) group.
Conclusions
The sensitivity and specificity of the MyHPV chip test are similar to the HC2. Detecting HPV-16/18 with an HPV DNA chip test, which is commonly used in many Asian countries, is useful in assessing the risk of high-grade cervical lesions. -
Citations
Citations to this article as recorded by- Human papilloma virus identification in ocular surface squamous neoplasia by p16 immunohistochemistry and DNA chip test
Tina Shrestha, Won Choi, Ga Eon Kim, Jee Myung Yang, Kyung Chul Yoon
Medicine.2019; 98(2): e13944. CrossRef - Comparison of the PANArray HPV Genotyping Chip Test with the Cobas 4800 HPV and Hybrid Capture 2 Tests for Detection of HPV in ASCUS Women
Eun Young Ki, Yoon Kyung Lee, Ahwon Lee, Jong Sup Park
Yonsei Medical Journal.2018; 59(5): 662. CrossRef
- Human papilloma virus identification in ocular surface squamous neoplasia by p16 immunohistochemistry and DNA chip test
- Difference of Genome-Wide Copy Number Alterations between High-Grade Squamous Intraepithelial Lesions and Squamous Cell Carcinomas of the Uterine Cervix
- Bum Hee Lee, Sangyoung Roh, Yu Im Kim, Ahwon Lee, Su Young Kim
- Korean J Pathol. 2012;46(2):123-130. Published online April 25, 2012
- DOI: https://doi.org/10.4132/KoreanJPathol.2012.46.2.123
- 8,481 View
- 46 Download
- 2 Crossref
-
 Abstract
Abstract
 PDF
PDF Background About 10% of high-grade squamous intraepithelial lesions (HSILs) progress to invasive carcinomas within 2-10 years. By delineating the events that occur in the early stage of the invasion, the pathogenesis of cervical cancer could be better understood. This will also propose the possible methods for inhibiting the tumor invasion and improving the survival of patients.
Methods We compared the genomic profiles between the HSIL and the invasive squamous cell carcinoma (SCC) using an array comparative genomic hybridization. Using recurrently altered genes, we performed a principal component analysis to see variation of samples in both groups. To find possibly affected pathways by altered genes, we analyzed genomic profiles with the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database and GOEAST software.
Results We found 11q12.3 and 2p24.1 regions have recurrent copy number gains in both groups. 16p12-13 and 20q11-13 regions showed an increased copy number only in cases of HSIL. 1q25.3 and 3q23-29 regions showed copy number gains only in cases of SCC. Altered genes in the SCC group were related to the mitogen-activated protein kinase signaling pathway and the RNA transport. Altered genes in the HSIL group were related to the ubiquitin mediated proteolysis and cell adhesion molecules.
Conclusions Our results showed not only that gains in 11q12.3 and 2p24.1 were early events occurring in the premalignant lesions and then maintained in cases of SCC but also that gains in 1q25.3 and 3q23-29 were late events occurring after invasion in those of SCC.
-
Citations
Citations to this article as recorded by- Cytokeratin and protein expression patterns in squamous cell carcinoma of the oral cavity provide evidence for two distinct pathogenetic pathways
GESCHE FROHWITTER, HORST BUERGER, PAUL J. VAN DIEST, EBERHARD KORSCHING, JOHANNES KLEINHEINZ, THOMAS FILLIES
Oncology Letters.2016; 12(1): 107. CrossRef - 'Drawing' a Molecular Portrait of CIN and Cervical Cancer: a Review of Genome-Wide Molecular Profiling Data
Olga V Kurmyshkina, Pavel I Kovchur, Tatyana O Volkova
Asian Pacific Journal of Cancer Prevention.2015; 16(11): 4477. CrossRef
- Cytokeratin and protein expression patterns in squamous cell carcinoma of the oral cavity provide evidence for two distinct pathogenetic pathways
- Prognostic Significance of Amplification of the c-MYC Gene in Surgically Treated Stage IB-IIB Cervical Cancer.
- Tae Jung Kim, Ahwon Lee, Sung Jong Lee, Won Chul Lee, Yeong Jin Choi, Kyo Young Lee, Chang Suk Kang
- Korean J Pathol. 2011;45(6):596-603.
- DOI: https://doi.org/10.4132/KoreanJPathol.2011.45.6.596
- 4,998 View
- 42 Download
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
Mutations of c-MYC have been described in cervical cancer. However, association between c-MYC gene status and its prognostic significance have not been clarified.
METHODS
Tissue microarray sections from 144 patients with stage IB-IIB cervical cancer treated by radical hysterectomy were analyzed by fluorescence in situ hybridization using a region-specific probe for c-MYC and a centromere-specific probe for chromosome 8.
RESULTS
Seventy five percent (108/144) of c-MYC gain and 6.9% (10/144) of c-MYC gene amplification were observed. c-MYC gene alteration was more frequently observed in squamous cell carcinoma than adenocarcinoma or adenosquamous carcinoma and were associated with low Ki67 labeling index (p=0.013). c-MYC amplification was not associated with clinicopathologic parameters except absence of bcl2 expression (p=0.048). Survival analysis revealed that patients with c-MYC amplification were significantly associated with higher risk of disease recurrence (p=0.007) and cancer related death (p=0.020). However, c-MYC gain was not associated with unfavorable outcome. Multivariate analysis proved c-MYC amplification as independent prognostic factors of shorter disease free survival and cancer-related death (p=0.028 and p=0.025, respectively).
CONCLUSIONS
c-MYC amplification, not gain, is an independent prognostic marker for shorter disease free and cancer specific survival in cervical cancer treated by radical hysterectomy. -
Citations
Citations to this article as recorded by- A Rare Case of Cutaneous Plasmacytosis in a Korean Male
Corey Georgesen, Meenal Kheterpal, Melissa Pulitzer
Case Reports in Pathology.2017; 2017: 1. CrossRef
- A Rare Case of Cutaneous Plasmacytosis in a Korean Male
- Detection Limit of Monoclonal B-Cells Using Multiplex PCR and Laser-Induced Fluorescence Capillary Electrophoresis.
- Sung Hak Lee, Yeonsook Moon, Byunghoo Song, Hyung Nam Lee, Ahwon Lee, Eun Sun Jung, Yeong Jin Choi, Kyo Young Lee, Chang Suk Kang, Gyeongsin Park
- Korean J Pathol. 2011;45(6):582-588.
- DOI: https://doi.org/10.4132/KoreanJPathol.2011.45.6.582
- 4,070 View
- 28 Download
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
The identification of monoclonality has been widely used for making diagnoses of lymphoproliferative lesions. Awareness of the sensitivity and detection limit of the technique used would be important for the data to be convincing.
METHODS
We investigated the minimum requirement of cells and sensitivity of gel electrophoresis (GE) and laser-induced fluorescence capillary electrophoresis (LFCE) for identifying IgH gene rearrangement using BIOMED-2 protocols. DNA extracted from Raji cells were diluted serially with peripheral blood mononuclear cells (PBMNCs) DNA. DNA from mixtures of diffuse large B-cell lymphoma (DLBCL) and reactive lymph nodes were also serially diluted.
RESULTS
For Raji cells, the detection limit was 62 and 16 cell-equivalents for GE and LFCE, respectively. In the condition with PBMNCs mixture, 2.5% and 1.25% of clonal cells was the minimum requirement for GE and LFCE, respectively. In 23% of DLBCL cells in tissue section, the detection limit was 120 and 12 cell-equivalents for GE and LFCE, respectively. In 3.2% of DLBCL cells, that was 1,200 and 120 cell-equivalents for GE and LFCE, respectively.
CONCLUSIONS
These results show that LFCE method is more sensitive than GE and the sensitivity of clonality detection can be influenced by the amount of admixed normal lymphoid cells. -
Citations
Citations to this article as recorded by- Molecular pathology diagnosis of diffuse large B cell lymphoma using BIOMED-2 clonal gene rearrangements
Saeid Ghorbian
Annals of Diagnostic Pathology.2017; 29: 28. CrossRef
- Molecular pathology diagnosis of diffuse large B cell lymphoma using BIOMED-2 clonal gene rearrangements
- The Usefulness of p16INK4a Immunocytochemical Staining in ASC-H Patients.
- Kwang Il Yim, Yeo Ju Kang, Tae Eun Kim, Gyeongsin Park, Eun Sun Jung, Yeong Jin Choi, Kyo Young Lee, Chang Seok Kang, Ahwon Lee
- Korean J Pathol. 2011;45(3):290-295.
- DOI: https://doi.org/10.4132/KoreanJPathol.2011.45.3.290
- 4,676 View
- 26 Download
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
The grey zone of cervical cytology, and in particular atypical squamous cells, cannot exclude HSIL (ASC-H) causes diagnostic difficulties and increases medical expenses. We analyzed p16INK4a expression in ASC-H liquid-based cytology specimens (LBCS) to develop more effective methods for the management of ASC-H patients.
METHODS
We carried out p16INK4a immunostaining with 57 LBCS of ASC-H diagnostic categories, all of which were histologically cofirmed and 43 cases of which were compared with the results of a human papillomavirus (HPV) chip test.
RESULTS
p16INK4a immunostaining with ASC-H LBCS was positive in 20% (3/15) of cervicitis, 25.0% (3/12) of tissue-low-grade squamous intraepithelial lesion, 75.0% (18/24) of tissue-high grade squamous intraepithelial lesion (HSIL), and 100% (6/6) of invasive cancer cases. The positivity of p16INK4a in LBCS was correlated with higher grade of histologic diagnosis (r=0.578, p=0.000). The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of p16INK4a immunostaining for the prediction of tissue-HSIL+ were 80.0%, 77.8%, 80.0%, and 77.8%, respectively. The sensitivity, specificity, PPV, and NPV of p16INK4a immunostaining plus HPV chip test for predicting tissue-HSIL+ were 71.2%, 86.4%, 84.2%, and 79.2%.
CONCLUSIONS
p16INK4a immunostaining as well as HPV chip testing with remaining LBCS with ASC-H are useful objective markers for the prediction of tissue-HSIL+. -
Citations
Citations to this article as recorded by- Usefulness of p16INK4a Immunocytochemical staining for the Differentiation between Atrophy and ASCUS in Diagnosis of Uterine Cervical Cancer
Hye Ryoung Shin, Taekil Eom, Wan-Su Choi
Biomedical Science Letters.2023; 29(3): 144. CrossRef
- Usefulness of p16INK4a Immunocytochemical staining for the Differentiation between Atrophy and ASCUS in Diagnosis of Uterine Cervical Cancer
- Copy Number Alterations of BCAS1 in Squamous Cell Carcinomas.
- Yu Im Kim, Ahwon Lee, Jennifer Kim, Bum Hee Lee, Sung Hak Lee, Suk Woo Nam, Sug Hyung Lee, Won Sang Park, Nam Jin Yoo, Jung Young Lee, Sang Ho Kim, Su Young Kim
- Korean J Pathol. 2011;45(3):271-275.
- DOI: https://doi.org/10.4132/KoreanJPathol.2011.45.3.271
- 4,482 View
- 34 Download
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
Breast carcinoma amplified sequence 1 (BCAS1), located in 20q13, is amplified and overexpressed in breast cancers. Even though BCAS1 is expected to be an oncogene candidate, its contribution to tumorigenesis and copy number status in other malignancies is not reported. To elucidate the role of BCAS1 in squamous cell carcinomas, we investigated the copy number status and expression level of BCAS1 in several squamous cell carcinoma cell lines, normal keratinocytes and primary tumors.
METHODS
We quantitated BCAS1 gene by real-time polymerase chain reaction (PCR). Expression level of BCAS1 was measured by real-time reverse transcription-PCR and immunoblot.
RESULTS
Seven (88%) of 8 squamous cell carcinoma cell lines showed copy number gain of BCAS1 with various degrees. BCAS1 gene in primary tumors (73%) also showed copy number gain. However, expression level did not show a linear correlation with copy number changes.
CONCLUSIONS
We identified copy number gain of BCAS1 in squamous cell carcinomas. Due to lack of linear correlation between copy numbers of BCAS1 and its expression level, we could not confirm that the overexpression of BCAS1 is a common finding in squamous cell carcinoma cell lines. However, this study shows that the copy number gain of BCAS1 is a common finding in squamous cell carcinomas. -
Citations
Citations to this article as recorded by- Electrochemical Approaches for Preparation of Tailor-Made Amino Acids
Nana Wang, Jingcheng Xu, Haibo Mei, Hiroki Moriwaki, Kunisuke Izawa, Vadim A. Soloshonok, Jianlin Han
Chinese Journal of Organic Chemistry.2021; 41(8): 3034. CrossRef
- Electrochemical Approaches for Preparation of Tailor-Made Amino Acids
- Evaluation of the HPV ISH Assay in Cervical Cancer.
- Jung Uee Lee, Jung Ha Shin, Jong Ok Kim, Yeong Jin Choi, Kyo Young Lee, Jong Sup Park, Won Chul Lee, Ahwon Lee
- Korean J Pathol. 2010;44(5):513-520.
- DOI: https://doi.org/10.4132/KoreanJPathol.2010.44.5.513
- 5,523 View
- 116 Download
- 3 Crossref
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
Human papillomavirus (HPV) infection can be detected by in situ hybridization (ISH), in which a punctate signal pattern indicates integrated HPV DNA and a diffuse pattern denotes the presence of episomal viral DNA. This study was conducted to evaluate the usefulness of an HPV ISH assay for invasive cervical cancer.
METHODS
The HPV ISH assay for high-risk HPV and immunohistochemical staining for p16(INK4a), p53, bcl-2, and Ki-67 were performed in a tissue microarray of 279 cervical cancers.
RESULTS
High-risk HPV ISH was positive in 194 (69.5%) of the samples. Punctate, diffuse, and mixed signal patterns were observed in 157 (56.3%), one (0.4%), and 36 cases (12.9%), respectively. Positive results in high-risk HPV ISH were associated with p16 and bcl-2 expression (p = 0.01 and p < 0.01, respectively). According to a Cox regression analysis, HPV infection and its surrogate immunohistochemical markers such as p16, bcl-2, and Ki-67 were not independent prognostic factors, but stage and grade were independent prognostic factors.
CONCLUSIONS
Our results confirm that an HPV ISH assay is reasonably sensitive for HPV infection and that it might be useful to identify integrated HPV DNA in formalin-fixed and paraffin-embedded specimens. Further study encompassing HPV type, E2/E6 ratio, and therapeutic modality is necessary to understand the clinical meaning of HPV status in cervical cancer. -
Citations
Citations to this article as recorded by- Prevalence of human papillomavirus in eyelid carcinoma among Koreans: a clinicopathological study
Min Kyu Yang, Namju Kim, Hokyung Choung, Ji Eun Kim, Sang In Khwarg
BMC Ophthalmology.2023;[Epub] CrossRef - Cervical cancer screening by molecular Pap‐transformation of gynecologic cytology
Shaikhali M Barodawala, Kirti Chadha, Vikas Kavishwar, Anuradha Murthy, Shamma Shetye
Diagnostic Cytopathology.2019; 47(5): 374. CrossRef - Prognostic Significance of Amplification of thec-MYCGene in Surgically Treated Stage IB-IIB Cervical Cancer
Tae-Jung Kim, Ahwon Lee, Sung-Jong Lee, Won-Chul Lee, Yeong-Jin Choi, Kyo-Young Lee, Chang Suk Kang
The Korean Journal of Pathology.2011; 45(6): 596. CrossRef
- Prevalence of human papillomavirus in eyelid carcinoma among Koreans: a clinicopathological study
- Comparison of Clinical Efficacy between an HPV DNA Chip and a Hybrid-Capture II Assay in a Patient with Abnormal Colposcopic Findings.
- Tae Jung Kim, Chan Kwon Jung, Ahwon Lee, Eun Sun Jung, Young Jin Choi, Kyo Young Lee, Jong Sup Park
- J Pathol Transl Med. 2008;19(2):119-125.
- DOI: https://doi.org/10.3338/kjc.2008.19.2.119
- 3,258 View
- 13 Download
- 1 Crossref
-
 Abstract
Abstract
 PDF
PDF - This study was performed to compare the efficacy between a DNA chip method and a Hybrid-Capture II assay (HC-II) for detecting human papillomavirus in patients with intraepithelial lesions of the uterine cervix. From May, 2005, to June, 2006, 192 patients with abnormal colposcopic findings received cervical cytology, HC-II and HPV DNA chip tests, and colposcopic biopsy or conization. We compared the results of HC-II and HPV DNA chip in conjunction with liquid based cervical cytology (LBCC) and confirmed the results of biopsy or conization. The sensitivity of the HPV DNA chip test was higher than HC-II or LBCC. The HPV DNA chip in conjunction with LBCC showed higher sensitivity than any single method and higher sensitivity than HC-II with LBCC. We confirmed that the HPV DNA chip test was more sensitive for detecting HPV in cervical lesions than HC-II, and that it would provide more useful clinical information about HPV type and its multiple infections.
-
Citations
Citations to this article as recorded by- Comparison of Analytical and Clinical Performance of HPV 9G DNA Chip, PANArray HPV Genotyping Chip, and Hybrid-Capture II Assay in Cervicovaginal Swabs
Ho Young Jung, Hye Seung Han, Hyo Bin Kim, Seo Young Oh, Sun-Joo Lee, Wook Youn Kim
Journal of Pathology and Translational Medicine.2016; 50(2): 138. CrossRef
- Comparison of Analytical and Clinical Performance of HPV 9G DNA Chip, PANArray HPV Genotyping Chip, and Hybrid-Capture II Assay in Cervicovaginal Swabs
- Epithelial-Myoepithelial Carcinoma of the Parotid Gland: Report of a Case Misinterpreted as Pleomorphic denoma on Fine Needle Aspiration Cytology.
- Dong Chul Kim, Ahwon Lee, Kyo Young Lee, Cang Suk Kang, Sang In Shim
- J Pathol Transl Med. 2002;13(1):42-46.
- 2,214 View
- 17 Download
-
 Abstract
Abstract
 PDF
PDF - Epithelial-myoepithelial carcinoma(EMC) is a rare, low grade malignant tumor of the salivary glands. The EMC has a distinctive histological appearance comprising ductal structures with an inner epithelial cell component and an outer layer of myoepithelial cells which show plump clear cytoplasm. The cytologic features of the EMC have been rarely described. A correct cytological diagnosis to this rare tumor is difficult with high false negative rate. We report a case of EMC in which fine needle aspiration cytologic findings were misinterpreted as a pleomorphic adenoma.
- Invasive Ductal Carcinoma Arising in a Recurrent Malignant Phyllodes Tumor: A Case Report.
- Ahwon Lee, Gyeongsin Park, Kyo Young Lee, Chang Suk Kang, Byung Kee Kim, Sang In Shim
- Korean J Pathol. 2005;39(2):134-136.
- 2,031 View
- 22 Download
-
 Abstract
Abstract
 PDF
PDF - We report here on a case of invasive ductal carcinoma arising in a recurrent malignant phyllodes tumor. The patient was a 33-year-old woman who presented with a left breast mass, and an excision was then performed. The mass, measuring 7.0 x 4.0 cm in size, was relatively well demarcated with a nodular contour and showed pale gray and solid cut surface with clefts on it. Histologically, the mass mainly consisted of stromal components that were characterized by high cellularity, marked nuclear atypism and brisk mitosis. The sparse glandular components were leaf-like in shape and lined by bland ductal epithelium without any nuclear atypism. Sixteen months later, the patient revisited our hospital with a recurrent mass, and underwent total mastectomy. The recurrent mass contained foci of definite invasive ductal carcinoma in the background of malignant phyllodes tumor, which was identical to the primary mass. This case demonstrates that it is possible that an invasive ductal carcinoma might arise within, at least with, a recurrent malignant phyllodes tumor.
- Fine Needle Aspiration Cytology of Mucinous Cystic Carcinoma of the Pancreas: A Case Report.
- Kyungji Lee, Ahwon Lee, Kyo Young Lee, Chang Suk Kang, Sang In Shim
- J Pathol Transl Med. 2005;16(2):88-92.
- 1,943 View
- 15 Download
-
 Abstract
Abstract
 PDF
PDF - Mucious cystic neoplasm of pancreas is a cystic neoplasm composed of columnar, mucin-producing epithelium and is supported by ovarian-type stroma. The key to the cytologic evaluation of pancreatic cystic lesions is to recognize the cytologic components as being diagnostic of a mucin-producing cystic neoplasm, as all of these neoplasms need to be resected. We report the use of fine needle aspiration cytology in the diagnosis of an invasive mucinous cystic carcinoma confirmed by partial pancreatectomy. The cytologic specimen showed a abundant mucin background and sheets or papillae of neoplastic cells. There are mucin-containing columnar cells that show a variable degree of cytologic atypia.
- Fine Needle Aspiration Cytology of the Warthin's Tumor Misinterpretated as Squamous Cell Carcinoma: A Case Report.
- Kyungji Lee, Chan Kwon Jung, Ahwon Lee, Kyo Young Lee, Chang Suk Kang
- J Pathol Transl Med. 2005;16(2):106-109.
- 4,020 View
- 109 Download
-
 Abstract
Abstract
 PDF
PDF - We report a case of Warthin's tumor of the parotid gland in a 53?year?old man, which is incorrectly diagnosed as squamous cell carcinoma. Fine needle aspiration cytology(FNAC) smear obtained from the right parotid gland revealed scattered epithelial cell clusters or nests in a diffuse inflammatory and necrotic background. Some epithelial cells had squamoid appearance showing variable sized bizarre shaped nuclei. They had abundant of dense eosinophilic keratinized cytoplasm. Occasionally, parakeratotic cells were also present. These cytologic findings with significant atypia and necrotic background made diagnosis as squamous cell carcinoma. But, the resection specimen from this patient showed classic Warthin's tumor in addition to abundant areas of inflammation and squamous metaplasia. Metaplastic or infarcted Warthin's tumor in the salivary gland may be confused with false positive diagnosis of malignancy on FNAC. Therefore, cytopathologist should have adequate awareness of potential of erroneous diagnosis in FNAC of Warthin's tumor.
- A Comparision of Surepath(TM) Liquid-Based Smear with a Conventional Smear for Cervicovaginal Cytology-with Reference to a Histological Diagnosis.
- Kyung Chul Lee, Chan Kwon Jung, Ahwon Lee, Eun Sun Jung, Yeong Jin Choi, Jong Sup Park, Kyo Young Lee
- J Pathol Transl Med. 2007;18(1):20-28.
- 2,724 View
- 47 Download
-
 Abstract
Abstract
 PDF
PDF - This study was performed to compare Surepath(TM) liquid-based smear and a conventional cervicovaginal smear with reference to a histological diagnosis. A hybrid capture test (HCII) was also performed and analyzed. We collected matched cases for cervicovaginal cytology- histology: 207 cases for conventional cytology (CC) and 199 cases for liquid-based cytology (LBC). HCII was performed in 254 patients. When a cytological diagnosis of ASCUS or above (ASCUS+) is classified as positive and a histological diagnosis of LSIL+ is classified as positive, the sensitivity and specificity for LBC was 91.7% and 75.9%, respectively and the sensitivity and specificity for CC was 62.6% and 96.1%, respectively. When a cytological and histological diagnosis of LSIL+ is classified as positive, the sensitivity and specificity for LBC was 77.5 and 96.6%, respectively and the sensitivity and specificity for CC was 49.7% and 100%, respectively. When a histological diagnosis of LSIL+ is classified as positive, the sensitivity and specificity for HCII was 78.9% and 78.1%, respectively. The concordance ratio between the cytological and histological diagnosis was 80.4% (kappa=76.0) for LBC and 56.5% (kappa=55.1) for CC. LBC is more sensitive and less specific then CC, as a cytological cutoff level of ASCUS, but more sensitive and equally specific, as a cytological cutoff level LSIL or HSIL. LBC is more reliable with a high concordance ratio between the cytological and histological diagnosis.
- Clinicopathologic Analysis of the Micropapillary Variant of Urothelial Carcinoma in Urinary.
- Kyungji Lee, Ahwon Lee, Yeong Jin Choi, Kyo Young Lee, Chang Suk Kang, Sang In Shim
- Korean J Pathol. 2006;40(4):263-268.
- 2,197 View
- 20 Download
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
Micropapillary urothelial carcinoma of urinary bladder is a rare and aggressive subtype of urothelial carcinoma (UC).
METHODS
AND RESULTS: Seven UCs with a micropapillary component (MPC) were identified by reviewing 135 cystectomy specimens of UC (5.2% in incidence). MPC was associated with conventional UC in 6 cases and the plasmacytoid variant of UC in 1 case. Lymph node metastasis, that characteristically contained MPC was present in 60% (3 out of 5 cases of regional lymph node dissection). Three patients with extensive MPC showed laminar propria invasion (pT1; 33%) and perivesical fat invasion (pT3; 67%). Two out of 3 patients with extensive MPC showed distant metastasis into the colon after cystectomy. The colonic lesions showed exclusively micropapillary differentiation. Four patients with focal or moderate MPC (pT2, 25%; pT3, 75%) were alive without disease at the time of writing this article. All 3 cases with extensive MPC had surface and/or invasive MPC on the prior TURB specimen. Immunohistochemically, the tumor cells were positive for cytokeratin 7, cytokeratin 20, EMA and E-cadherin and tissue retraction spaces that simulate lymphatic spaces were negative for CD34 in all 7 cases.
CONCLUSIONS
This study suggests that the micropapillary growth pattern in UC is a manifestation of aggressive behavior and UC with MPC must be included as part of the differential diagnosis when dealing with a metastatic lesion with a micropaillary structure.
- Restrictive Dermopathy In Two Siblings.
- Tae Jung Kim, Youn Soo Lee, Hyun Young Ahn, Ahwon Lee, Kyo Young Lee, Jong Sup Park
- Korean J Pathol. 2007;41(1):47-50.
- 1,980 View
- 18 Download
-
 Abstract
Abstract
 PDF
PDF - Restrictive dermopathy (RD) is a rare and lethal autosomal recessive skin disorder that presents with rigid and tense skin, multiple joint contractures and fixed facial expression. We report herein two siblings from consecutive pregnancies affected with RD. Both siblings died of respiratory insufficiency at a day after birth. An autopsy for the first baby and a skin necropsy for the second baby were performed. The gross findings of both were characterized by thin, translucent skin with prominent vessels, multiple joint contractures resulting in hyperflexed position, and a typical facial appearance with a fixed open mouth in the O-position. Such manifestations are typical features of RD. At the autopsy of the first baby, no internal organ abnormality was found. The histologic findings of the skin of the second baby revealed a thin dermis consisting of a flat dermal-epidermal junction, hypoplastic skin appendages and compactly arranged collagen bundles. Elastic tissue stain showed markedly decreased elastic fibers.
- Prognostic Significance of P53, BCL-2 and PCNA in Diffuse Large B-Cell Lymphoma: Correlation with International Prognostic Index.
- Dong chul Kim, Gyeongsin Park, Ahwon Lee, Kyo Young Lee, Sang In Shim, Chang Suk Kang
- Korean J Pathol. 2003;37(6):407-412.
- 1,912 View
- 22 Download
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
Diffuse large B-cell lymphoma (DLBCL) represents a diverse spectrum of clinical presentation, morphology, and genetic and molecular alterations, and shows variable prognoses and responses to therapy. The International Prognosis Index (IPI) is widely used to predict prognosis but is not precise.
METHODS
Thirty-nine cases of DLBCL were classified into low- and high-risk groups according to IPI and were analyzed for their p53, BCL-2, BCL-6 and PCNA expression profile by immunohistochemical staining and overall survival rate.
RESULTS
The mean age of the 39 patients, 23 males and 16 females, was 52.6 years. There were 23 cases (59.0%) in the low-risk group and 16 (41.0%) in the high-risk group. p53, BCL-2, BCL-6 and PCNA expression was higher in the high-risk group than in the low-risk group, but only the differences in p53 and BCL-2 expression were statistically significant (p < 0.05).
CONCLUSION
The p53 and BCL-2 protein expression in DLBCL may supplement IPI in predicting the prognosis of DLBCL patients.
- Primary Diffuse Large B-cell Lymphoma of the Prostate: A Case Report.
- Dong chul Kim, Gyeongsin Park, Ahwon Lee, Kyungja Han, Chang Suk Kang
- Korean J Pathol. 2003;37(6):432-434.
- 2,235 View
- 20 Download
-
 Abstract
Abstract
 PDF
PDF - Primary lymphomas of the prostate are extremely rare and can mimic other more common prostatic lesions clinically. We report a case of primary diffuse large B-cell lymphoma of the prostate in an 81-year-old man. The patient presented with voiding difficulty as an initial symptom and enlargement of the prostate on rectal digital examination. Transurethral prostatic resection was performed. On microscopic examination, atypical lymphoid cells infiltrated and replaced the prostatic parenchyma. The tumor cells had large nuclei with irregular nuclear membrane and vesicular clumped chromatin. Nucleoli were not distinct and the cells had scanty cytoplasm. Immunohistochemically, the tumor cells were immunoreactive for CD20 and CD79a but not reactive for CD5, BCL-2 and BCL-6. Histopathological diagnosis was diffuse large B-cell lymphoma of the prostate. The patient received 5 cycles of chemotherapy after histologic diagnosis but died from pulmonary and scrotal metastases 6 months later.
- The Expression of Telomerase Reverse Transcriptase Protein is an Independent Prognostic Marker in Early Stage Non-Small Cell Lung Carcinomas.
- Ji Han Jung, Chan Kwon Jung, Ahwon Lee, Gyeongsin Park, Jinyoung Yoo, Kyo Young Lee
- Korean J Pathol. 2007;41(2):95-102.
- 2,294 View
- 18 Download
-
 Abstract
Abstract
 PDF
PDF - BACKGROUND
The catalytic subunit of telomerase, hTERT (telomerase reverse transcriptase), is one of the most important components of telomerase, and performs a pivotal role in the mechanism underlying the regulation of telomerase activity in cellular immortalization and carcinogenesis. The principal objective of this study was to investigate hTERT expression in patients with non-small cell lung carcinomas (NSCLCs), and to evaluate its clinical significance and association with the expression of p16 and p53.
METHODS
Using tissue microarray, the protein expression profiles of hTERT, p16 and p53 were investigated via immunohistochemistry in 167 samples of NSCLCs.
RESULTS
Expression was observed in 54.5% (91/167) of the tumors, which were predominantly squamous cell carcinomas. Patients evidencing hTERT expression in their tumors exhibited significantly poorer survival rates than did patients without hTERT expression in early-stage NSCLCs (p=0.0125). According to the results of our Cox regression analysis, hTERT expression proved to be an independent prognostic factor (p=0.006), particularly for squamous cell carcinomas (p=0.019). hTERT expression was not correlated with p16 expression, but was rather associated with the expression of p53 (p=0.002).
CONCLUSIONS
Our results show that hTERT may perform a function in the progression of NSCLC, and that its detection may be useful in predicting the prognosis of NSCLC patients in the early stages of the disease, as well as in the development of a targeted therapy in these tumors.

 E-submission
E-submission







 First
First Prev
Prev



