Articles
- Page Path
- HOME > J Pathol Transl Med > Volume 54(1); 2020 > Article
-
Review
Standardized Pathology Report for Colorectal Cancer, 2nd Edition -
Baek-hui Kim1
 , Joon Mee Kim,2
, Joon Mee Kim,2 , Gyeong Hoon Kang3
, Gyeong Hoon Kang3 , Hee Jin Chang4
, Hee Jin Chang4 , Dong Wook Kang5
, Dong Wook Kang5 , Jung Ho Kim3
, Jung Ho Kim3 , Jeong Mo Bae3
, Jeong Mo Bae3 , An Na Seo6
, An Na Seo6 , Ho Sung Park7
, Ho Sung Park7 , Yun Kyung Kang8
, Yun Kyung Kang8 , Kyung-Hwa Lee9
, Kyung-Hwa Lee9 , Mee Yon Cho10
, Mee Yon Cho10 , In-Gu Do11
, In-Gu Do11 , Hye Seung Lee12
, Hye Seung Lee12 , Hee Kyung Chang13
, Hee Kyung Chang13 , Do Youn Park14
, Do Youn Park14 , Hyo Jeong Kang15
, Hyo Jeong Kang15 , Jin Hee Sohn11
, Jin Hee Sohn11 , Mee Soo Chang16
, Mee Soo Chang16 , Eun Sun Jung17
, Eun Sun Jung17 , So-Young Jin18
, So-Young Jin18 , Eunsil Yu15
, Eunsil Yu15 , Hye Seung Han19
, Hye Seung Han19 , Youn Wha Kim20
, Youn Wha Kim20 , The Gastrointestinal Pathology Study Group of the Korean Society of Pathologists
, The Gastrointestinal Pathology Study Group of the Korean Society of Pathologists -
Journal of Pathology and Translational Medicine 2020;54(1):1-19.
DOI: https://doi.org/10.4132/jptm.2019.09.28
Published online: November 13, 2019
1Department of Pathology, Korea University Guro Hospital, Seoul, Korea
2Department of Pathology, Inha University School of Medicine, Incheon, Korea
3Department of Pathology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
4Department of Pathology, Research Institute and Hospital, National Cancer Center, Goyang, Korea
5Department of Pathology, Eulji University Hospital, Eulji University School of Medicine, Daejeon, Korea
6Department of Pathology, School of Medicine, Kyungpook National University, Daegu, Korea
7Department of Pathology, Chonbuk National University Medical School, Jeonju, Korea
8Department of Pathology, Seoul Paik Hospital, Inje University College of Medicine, Seoul, Korea
9Department of Pathology, Chonnam National University Medical School, Gwangju, Korea
10Department of Pathology, Yonsei University Wonju College of Medicine, Wonju, Korea
11Department of Pathology, Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, Korea
12Department of Pathology, Seoul National University, Bundang Hospital, Seongnam, Korea
13Department of Pathology, Kosin University College of Medicine, Busan, Korea
14Department of Pathology, Pusan National University Hospital, Pusan National University School of Medicine, Busan, Korea
15Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
16Department of Pathology, Seoul National University Boramae Hospital, Seoul National University College of Medicine, Seoul, Korea
17Department of Hospital Pathology, College of Medicine, The Catholic University of Korea, Seoul, Korea
18Department of Pathology, Soonchunhyang University Seoul Hospital, Soonchunhyang UniversityCollege of Medicine, Seoul, Korea
19Department of Pathology, Konkuk University Medical Center, Konkuk University School of Medicine, Seoul, Korea
20Department of Pathology, Kyung Hee University College of Medicine, Seoul, Korea
- Corresponding Author: Joon Mee Kim, MD, PhD, Department of Pathology, Inha University School of Medicine, 27 Inhang-ro, Jung-gu, Incheon 22332, Korea Tel: +82-32-890-3987, Fax: +82-32-890-3464, E-mail: jmkpath@inha.ac.kr
© 2020 The Korean Society of Pathologists/The Korean Society for Cytopathology
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Figure & Data
References
Citations

- Diagnostic accuracy and pitfalls of MRI for restaging locally advanced rectal cancer in patients following anti-PD1 therapy plus neoadjuvant chemoradiotherapy: a multicenter study
Lixue Xu, Liting Sun, Yuhuan Fu, Guangyong Chen, Hongwei Yao, Zhenchang Wang, Zhenghan Yang, Jie Zhang
Abdominal Radiology.2025;[Epub] CrossRef - Unraveling the role of perineural invasion in cancer progression across multiple tumor types
Muqtada Shaikh, Sanket Shirodkar, Gaurav Doshi
Medical Oncology.2025;[Epub] CrossRef - Malt lymphoma affecting the oral cavity: a clinical, pathologic and genetic study of malt 1 gene translocation
Juan Manuel Arteaga Legarrea, Mauro Lima dos Santos, Nathalia Gomes Rodrigues, Ricardo Santiago Gomez, Ricardo Alves Mesquita, Silvia Ferreira de Sousa, Cinthia Verónica Bardález López de Cáceres, Hélder Antônio Rebelo Pontes, Pablo Agustin Vargas, Luiz A
Oral Surgery, Oral Medicine, Oral Pathology and Oral Radiology.2025;[Epub] CrossRef - Proteogenomic profiling predicts outcomes of adjuvant chemotherapy in extrahepatic cholangiocarcinoma
Hyehyun Jeong, Ji-Hye Oh, Hee-Sung Ahn, Baek-Yeol Ryoo, Kyu-pyo Kim, Jae Ho Jeong, Inkeun Park, Dae Wook Hwang, Jae Hoon Lee, Ki Byung Song, Woohyung Lee, Ki-Hun Kim, Deog-Bog Moon, Gi Won Song, Dong-Hwan Jung, Seung-Mo Hong, Chae Won Park, In-Pyo Baek, Y
Journal of Hepatology.2025;[Epub] CrossRef - Additional staining for lymphovascular invasion is associated with increased estimation of lymph node metastasis in patients with T1 colorectal cancer: Systematic review and meta‐analysis
Jun Watanabe, Katsuro Ichimasa, Yuki Kataoka, Atsushi Miki, Hidehiro Someko, Munenori Honda, Makiko Tahara, Takeshi Yamashina, Khay Guan Yeoh, Shigeo Kawai, Kazuhiko Kotani, Naohiro Sata
Digestive Endoscopy.2024; 36(5): 533. CrossRef - The use of core descriptors from the ENiGMA code study in recent literature: a systematic review
Saher‐Zahra Khan, Andrea Arline, Kate M. Williams, Matthew J. Lee, Emily Steinhagen, Sharon L. Stein
Colorectal Disease.2024; 26(3): 428. CrossRef - Efficacy and safety of PD-1 blockade plus long-course chemoradiotherapy in locally advanced rectal cancer (NECTAR): a multi-center phase 2 study
Zhengyang Yang, Jiale Gao, Jianyong Zheng, Jiagang Han, Ang Li, Gang Liu, Yi Sun, Jie Zhang, Guangyong Chen, Rui Xu, Xiao Zhang, Yishan Liu, Zhigang Bai, Wei Deng, Wei He, Hongwei Yao, Zhongtao Zhang
Signal Transduction and Targeted Therapy.2024;[Epub] CrossRef - Diagnostic Accuracy of Highest-Grade or Predominant Histological Differentiation of T1 Colorectal Cancer in Predicting Lymph Node Metastasis: A Systematic Review and Meta-Analysis
Jun Watanabe, Katsuro Ichimasa, Yuki Kataoka, Shoko Miyahara, Atsushi Miki, Khay Guan Yeoh, Shigeo Kawai, Fernando Martínez de Juan, Isidro Machado, Kazuhiko Kotani, Naohiro Sata
Clinical and Translational Gastroenterology.2024; 15(3): e00673. CrossRef - Comparative evaluation of CT and MRI in the preoperative staging of colon cancer
Effrosyni Bompou, Aikaterini Vassiou, Ioannis Baloyiannis, Konstantinos Perivoliotis, Ioannis Fezoulidis, George Tzovaras
Scientific Reports.2024;[Epub] CrossRef - Pathologic Implications of Magnetic Resonance Imaging-detected Extramural Venous Invasion of Rectal Cancer
Hyun Gu Lee, Chan Wook Kim, Jong Keon Jang, Seong Ho Park, Young Il Kim, Jong Lyul Lee, Yong Sik Yoon, In Ja Park, Seok-Byung Lim, Chang Sik Yu, Jin Cheon Kim
Clinical Colorectal Cancer.2023; 22(1): 129. CrossRef - A standardized pathology report for gastric cancer: 2nd edition
Young Soo Park, Myeong-Cherl Kook, Baek-hui Kim, Hye Seung Lee, Dong-Wook Kang, Mi-Jin Gu, Ok Ran Shin, Younghee Choi, Wonae Lee, Hyunki Kim, In Hye Song, Kyoung-Mee Kim, Hee Sung Kim, Guhyun Kang, Do Youn Park, So-Young Jin, Joon Mee Kim, Yoon Jung Choi,
Journal of Pathology and Translational Medicine.2023; 57(1): 1. CrossRef - IGFL2-AS1, a Long Non-Coding RNA, Is Associated with Radioresistance in Colorectal Cancer
Jeeyong Lee, Da Yeon Kim, Younjoo Kim, Ui Sup Shin, Kwang Seok Kim, Eun Ju Kim
International Journal of Molecular Sciences.2023; 24(2): 978. CrossRef - A Standardized Pathology Report for Gastric Cancer: 2nd Edition
Young Soo Park, Myeong-Cherl Kook, Baek-hui Kim, Hye Seung Lee, Dong-Wook Kang, Mi-Jin Gu, Ok Ran Shin, Younghee Choi, Wonae Lee, Hyunki Kim, In Hye Song, Kyoung-Mee Kim, Hee Sung Kim, Guhyun Kang, Do Youn Park, So-Young Jin, Joon Mee Kim, Yoon Jung Choi,
Journal of Gastric Cancer.2023; 23(1): 107. CrossRef - Incremental Detection Rate of Dysplasia and Sessile Serrated Polyps/Adenomas Using Narrow-Band Imaging and Dye Spray Chromoendoscopy in Addition to High-Definition Endoscopy in Patients with Long-Standing Extensive Ulcerative Colitis: Segmental Tandem End
Ji Eun Kim, Chang Wan Choi, Sung Noh Hong, Joo Hye Song, Eun Ran Kim, Dong Kyung Chang, Young-Ho Kim
Diagnostics.2023; 13(3): 516. CrossRef - Prognostic Impact of Extramural Lymphatic, Vascular, and Perineural Invasion in Stage II Colon Cancer: A Comparison With Intramural Invasion
Sang Sik Cho, Ji Won Park, Gyeong Hoon Kang, Jung Ho Kim, Jeong Mo Bae, Sae-Won Han, Tae-You Kim, Min Jung Kim, Seung-Bum Ryoo, Seung-Yong Jeong, Kyu Joo Park
Diseases of the Colon & Rectum.2023; 66(3): 366. CrossRef - Towards targeted colorectal cancer biopsy based on tissue morphology assessment by compression optical coherence elastography
Anton A. Plekhanov, Marina A. Sirotkina, Ekaterina V. Gubarkova, Elena B. Kiseleva, Alexander A. Sovetsky, Maria M. Karabut, Vladimir E. Zagainov, Sergey S. Kuznetsov, Anna V. Maslennikova, Elena V. Zagaynova, Vladimir Y. Zaitsev, Natalia D. Gladkova
Frontiers in Oncology.2023;[Epub] CrossRef - Is High-Grade Tumor Budding an Independent Prognostic Factor in Stage II Colon Cancer?
Jung Kyong Shin, Yoon Ah Park, Jung Wook Huh, Seong Hyeon Yun, Hee Cheol Kim, Woo Yong Lee, Seok Hyung Kim, Sang Yun Ha, Yong Beom Cho
Diseases of the Colon & Rectum.2023; 66(8): e801. CrossRef - Detection of Human cytomegalovirus UL55 Gene and IE/E Protein Expression in Colorectal Cancer Patients in Egypt
May Raouf, Ahmed A. Sabry, Mahinour A. Ragab, Samar El Achy, Amira Amer
BMC Cancer.2023;[Epub] CrossRef - Polo-like kinase 4 as a potential predictive biomarker of chemoradioresistance in locally advanced rectal cancer
Hyunseung Oh, Soon Gu Kim, Sung Uk Bae, Sang Jun Byun, Shin Kim, Jae-Ho Lee, Ilseon Hwang, Sun Young Kwon, Hye Won Lee
Journal of Pathology and Translational Medicine.2022; 56(1): 40. CrossRef - A Prediction Model for Tumor Recurrence in Stage II–III Colorectal Cancer Patients: From a Machine Learning Model to Genomic Profiling
Po-Chuan Chen, Yu-Min Yeh, Bo-Wen Lin, Ren-Hao Chan, Pei-Fang Su, Yi-Chia Liu, Chung-Ta Lee, Shang-Hung Chen, Peng-Chan Lin
Biomedicines.2022; 10(2): 340. CrossRef - Rationale and design of a prospective, multicenter, phase II clinical trial of safety and efficacy evaluation of long course neoadjuvant chemoradiotherapy plus tislelizumab followed by total mesorectal excision for locally advanced rectal cancer (NCRT-PD1
Zhengyang Yang, Xiao Zhang, Jie Zhang, Jiale Gao, Zhigang Bai, Wei Deng, Guangyong Chen, Yongbo An, Yishan Liu, Qi Wei, Jiagang Han, Ang Li, Gang Liu, Yi Sun, Dalu Kong, Hongwei Yao, Zhongtao Zhang
BMC Cancer.2022;[Epub] CrossRef - Potential of DEK proto‑oncogene as a prognostic biomarker for colorectal cancer: An evidence‑based review
Muhammad Habiburrahman, Muhammad Wardoyo, Stefanus Sutopo, Nur Rahadiani
Molecular and Clinical Oncology.2022;[Epub] CrossRef - Reproducibility and Feasibility of Classification and National Guidelines for Histological Diagnosis of Canine Mammary Gland Tumours: A Multi-Institutional Ring Study
Serenella Papparella, Maria Crescio, Valeria Baldassarre, Barbara Brunetti, Giovanni Burrai, Cristiano Cocumelli, Valeria Grieco, Selina Iussich, Lorella Maniscalco, Francesca Mariotti, Francesca Millanta, Orlando Paciello, Roberta Rasotto, Mariarita Roma
Veterinary Sciences.2022; 9(7): 357. CrossRef - Composite scoring system and optimal tumor budding cut-off number for estimating lymph node metastasis in submucosal colorectal cancer
Jeong-ki Kim, Ye-Young Rhee, Jeong Mo Bae, Jung Ho Kim, Seong-Joon Koh, Hyun Jung Lee, Jong Pil Im, Min Jung Kim, Seung-Bum Ryoo, Seung-Yong Jeong, Kyu Joo Park, Ji Won Park, Gyeong Hoon Kang
BMC Cancer.2022;[Epub] CrossRef - Automated Hybrid Model for Detecting Perineural Invasion in the Histology of Colorectal Cancer
Jiyoon Jung, Eunsu Kim, Hyeseong Lee, Sung Hak Lee, Sangjeong Ahn
Applied Sciences.2022; 12(18): 9159. CrossRef - Clinical Implication of Perineural and Lymphovascular Invasion in Rectal Cancer Patients Who Underwent Surgery After Preoperative Chemoradiotherapy
Young Il Kim, Chan Wook Kim, Jong Hoon Kim, Jihun Kim, Jun-Soo Ro, Jong Lyul Lee, Yong Sik Yoon, In Ja Park, Seok-Byung Lim, Chang Sik Yu, Jin Cheon Kim
Diseases of the Colon & Rectum.2022; 65(11): 1325. CrossRef - Molecular Pathology of Gastric Cancer
Moonsik Kim, An Na Seo
Journal of Gastric Cancer.2022; 22(4): 264. CrossRef - Selective approach to arterial ligation in radical sigmoid colon cancer surgery with D3 lymph node dissection: A multicenter comparative study
Sergey Efetov, Albina Zubayraeva, Cüneyt Kayaalp, Alisa Minenkova, Yusuf Bağ, Aftandil Alekberzade, Petr Tsarkov
Turkish Journal of Surgery.2022; 38(4): 382. CrossRef - Evaluation of lncRNA FOXD2-AS1 Expression as a Diagnostic Biomarker in Colorectal Cancer
Hooman Shalmashi, Sahar Safaei, Dariush Shanehbandi, Milad Asadi, Soghra Bornehdeli, Abdolreza Mehdi Navaz
Reports of Biochemistry and Molecular Biology.2022; 11(3): 471. CrossRef - Improvement in the Assessment of Response to Preoperative Chemoradiotherapy for Rectal Cancer Using Magnetic Resonance Imaging and a Multigene Biomarker
Eunhae Cho, Sung Woo Jung, In Ja Park, Jong Keon Jang, Seong Ho Park, Seung-Mo Hong, Jong Lyul Lee, Chan Wook Kim, Yong Sik Yoon, Seok-Byung Lim, Chang Sik Yu, Jin Cheon Kim
Cancers.2021; 13(14): 3480. CrossRef - Addition of V-Stage to Conventional TNM Staging to Create the TNVM Staging System for Accurate Prediction of Prognosis in Colon Cancer: A Multi-Institutional Retrospective Cohort Study
Jung Hoon Bae, Ji Hoon Kim, Jaeim Lee, Bong-Hyeon Kye, Sang Chul Lee, In Kyu Lee, Won Kyung Kang, Hyeon-Min Cho, Yoon Suk Lee
Biomedicines.2021; 9(8): 888. CrossRef - Gene Expression Profiles Associated with Radio-Responsiveness in Locally Advanced Rectal Cancer
Jeeyong Lee, Junhye Kwon, DaYeon Kim, Misun Park, KwangSeok Kim, InHwa Bae, Hyunkyung Kim, JoonSeog Kong, Younjoo Kim, UiSup Shin, EunJu Kim
Biology.2021; 10(6): 500. CrossRef - A Patient-Derived Organoid-Based Radiosensitivity Model for the Prediction of Radiation Responses in Patients with Rectal Cancer
Misun Park, Junhye Kwon, Joonseog Kong, Sun Mi Moon, Sangsik Cho, Ki Young Yang, Won Il Jang, Mi Sook Kim, Younjoo Kim, Ui Sup Shin
Cancers.2021; 13(15): 3760. CrossRef - Comparison between Local Excision and Radical Resection for the Treatment of Rectal Cancer in ypT0-1 Patients: An Analysis of the Clinicopathological Factors and Survival Rates
Soo Young Oh, In Ja Park, Young IL Kim, Jong-Lyul Lee, Chan Wook Kim, Yong Sik Yoon, Seok-Byung Lim, Chang Sik Yu, Jin Cheon Kim
Cancers.2021; 13(19): 4823. CrossRef - Comparison of Vascular Invasion With Lymph Node Metastasis as a Prognostic Factor in Stage I-III Colon Cancer: An Observational Cohort Study
Jung Hoon Bae, Ji Hoon Kim, Bong-Hyeon Kye, Abdullah Al-Sawat, Chul Seung Lee, Seung-Rim Han, In Kyu Lee, Sung Hak Lee, Yoon Suk Lee
Frontiers in Surgery.2021;[Epub] CrossRef - Clinicopathological significance of Ki67 expression in colorectal cancer
Jing Li, Zhi-ye Liu, Hai-bo Yu, Qing Xue, Wen-jie He, Hai-tao Yu
Medicine.2020; 99(20): e20136. CrossRef - Lateral lymph node and its association with distant recurrence in rectal cancer: A clue of systemic disease
Young Il Kim, Jong Keon Jang, In Ja Park, Seong Ho Park, Jong Beom Kim, Jin-Hong Park, Tae Won Kim, Jun-Soo Ro, Seok-Byung Lim, Chang Sik Yu, Jin Cheon Kim
Surgical Oncology.2020; 35: 174. CrossRef - Transformation of Pathology Reports Into the Common Data Model With Oncology Module: Use Case for Colon Cancer
Borim Ryu, Eunsil Yoon, Seok Kim, Sejoon Lee, Hyunyoung Baek, Soyoung Yi, Hee Young Na, Ji-Won Kim, Rong-Min Baek, Hee Hwang, Sooyoung Yoo
Journal of Medical Internet Research.2020; 22(12): e18526. CrossRef
 PubReader
PubReader ePub Link
ePub Link-
 Cite this Article
Cite this Article
- Cite this Article
-
- Close
- Download Citation
- Close
- Figure






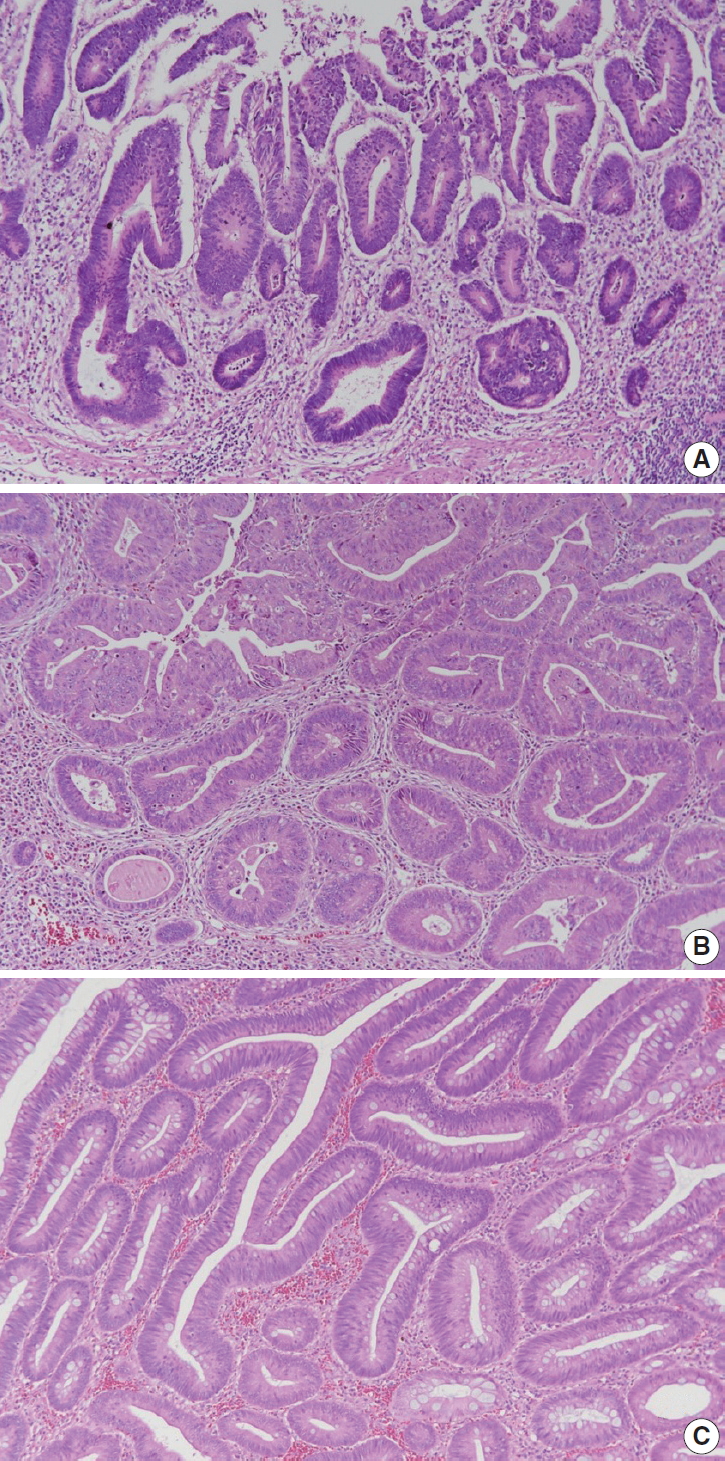
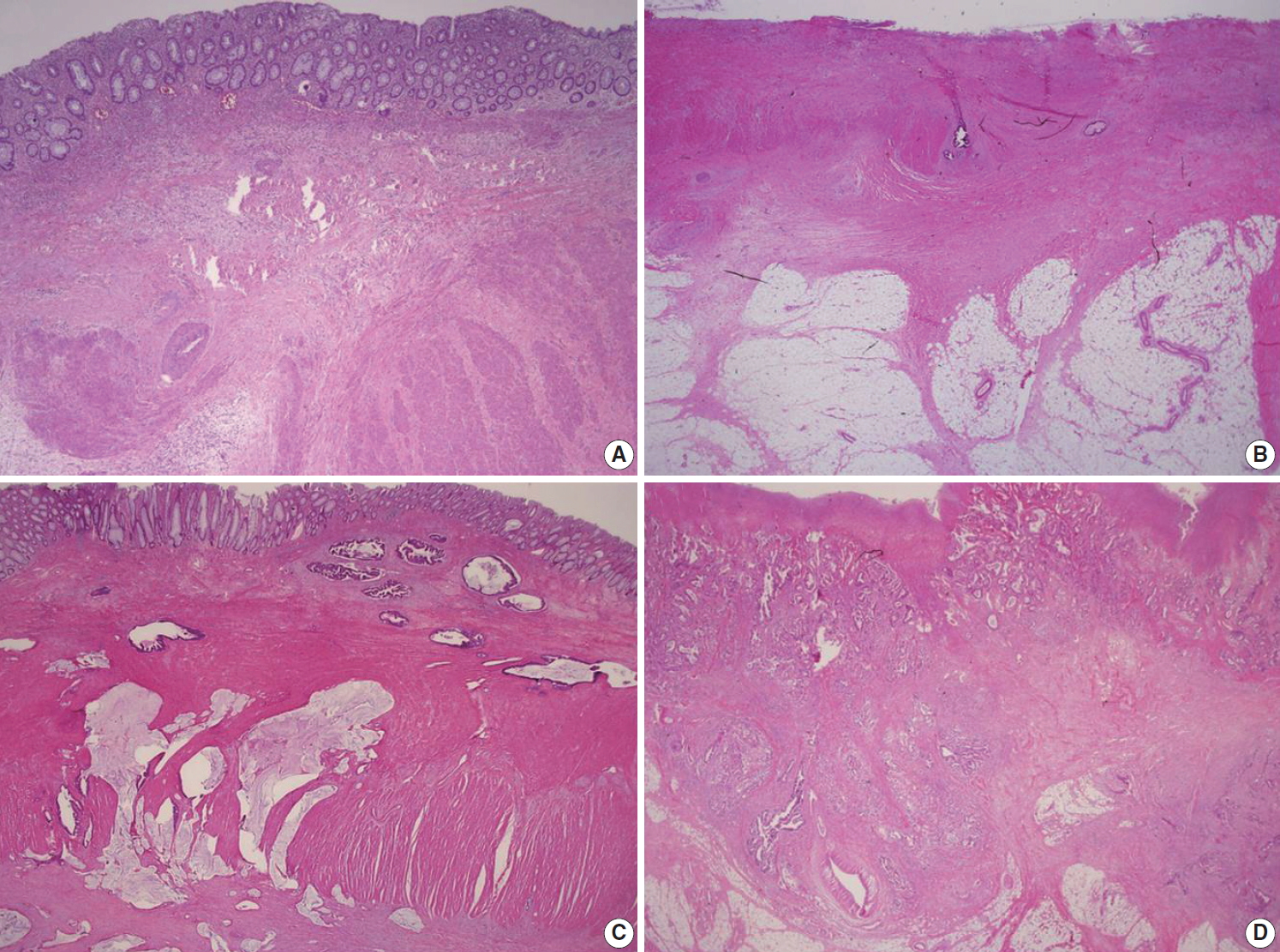
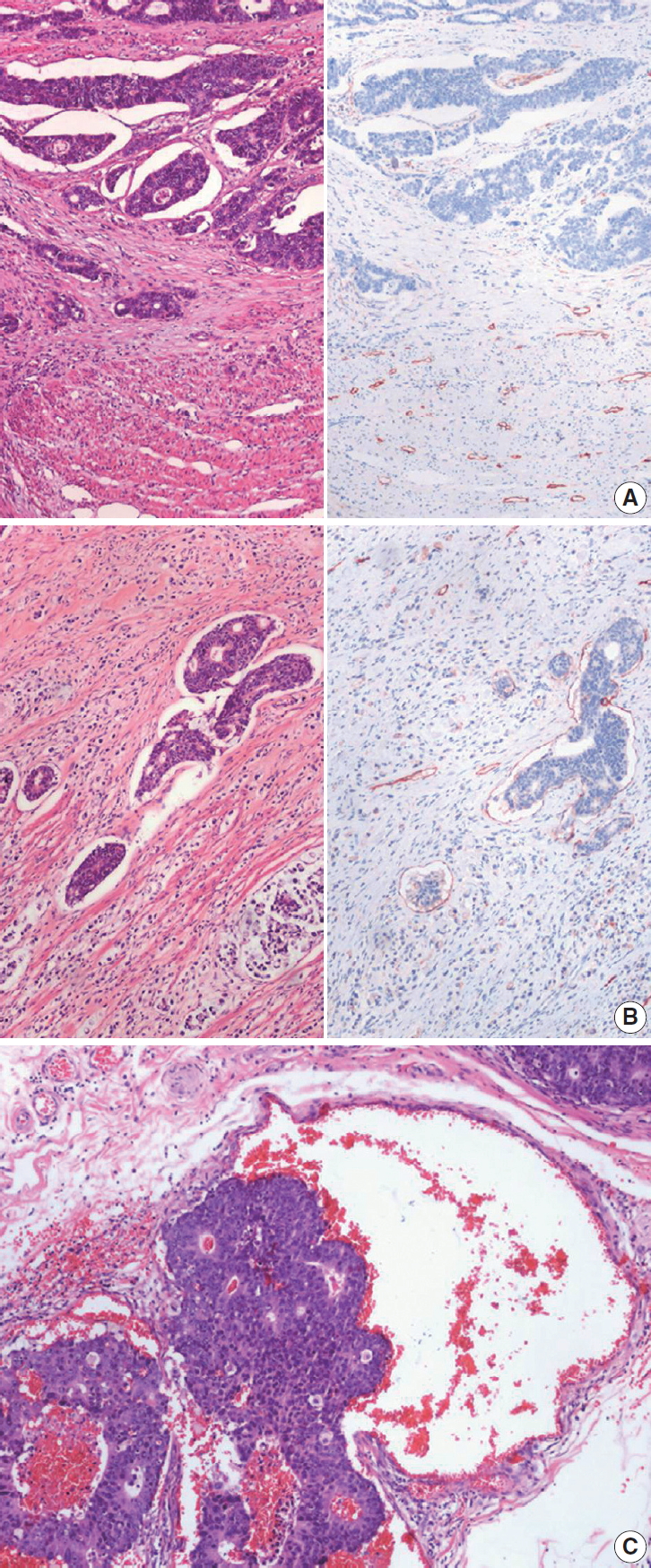
Fig. 1.
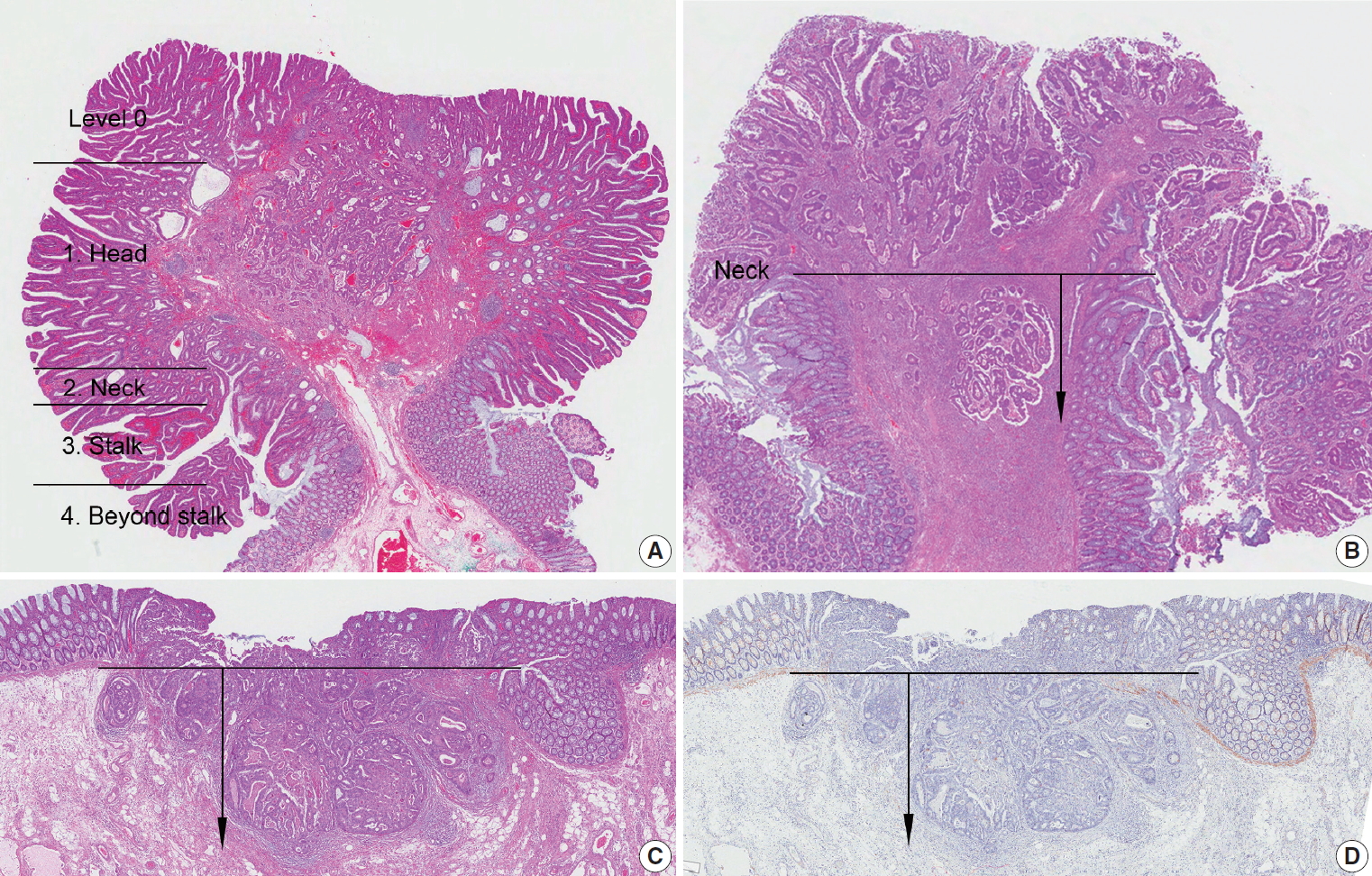
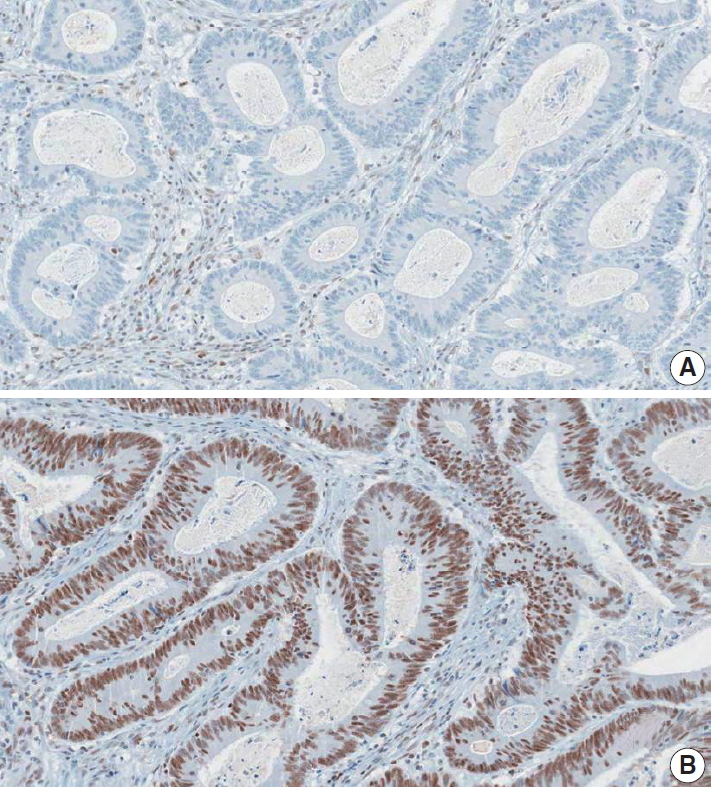
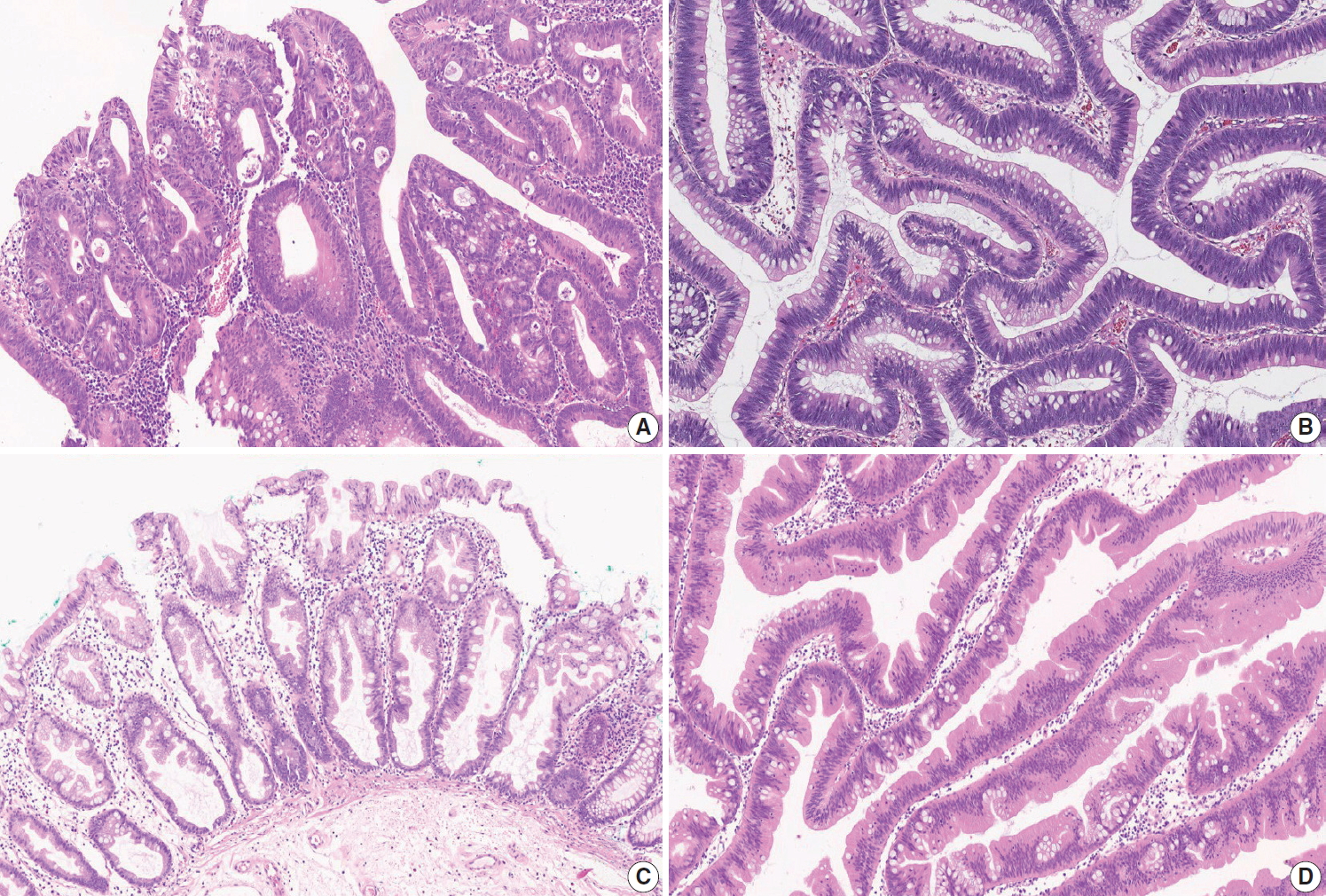
Fig. 2.
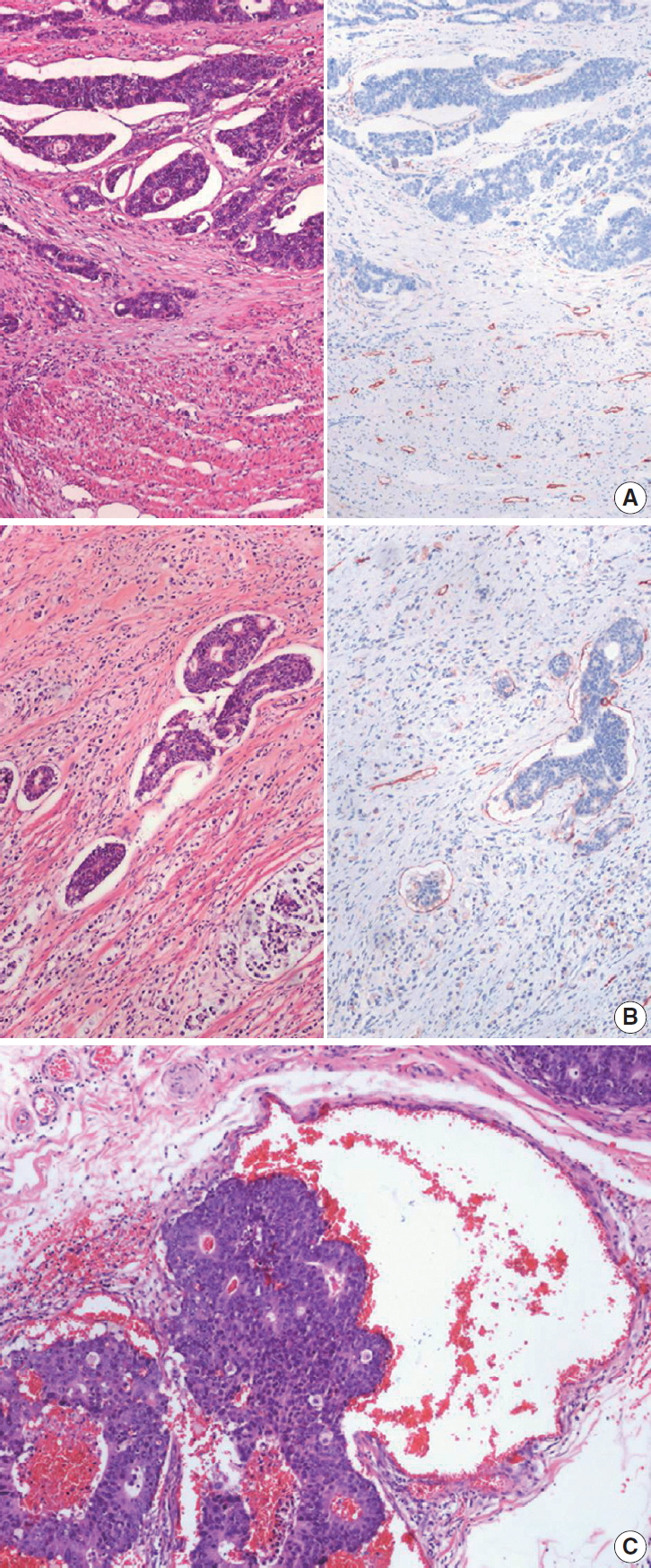
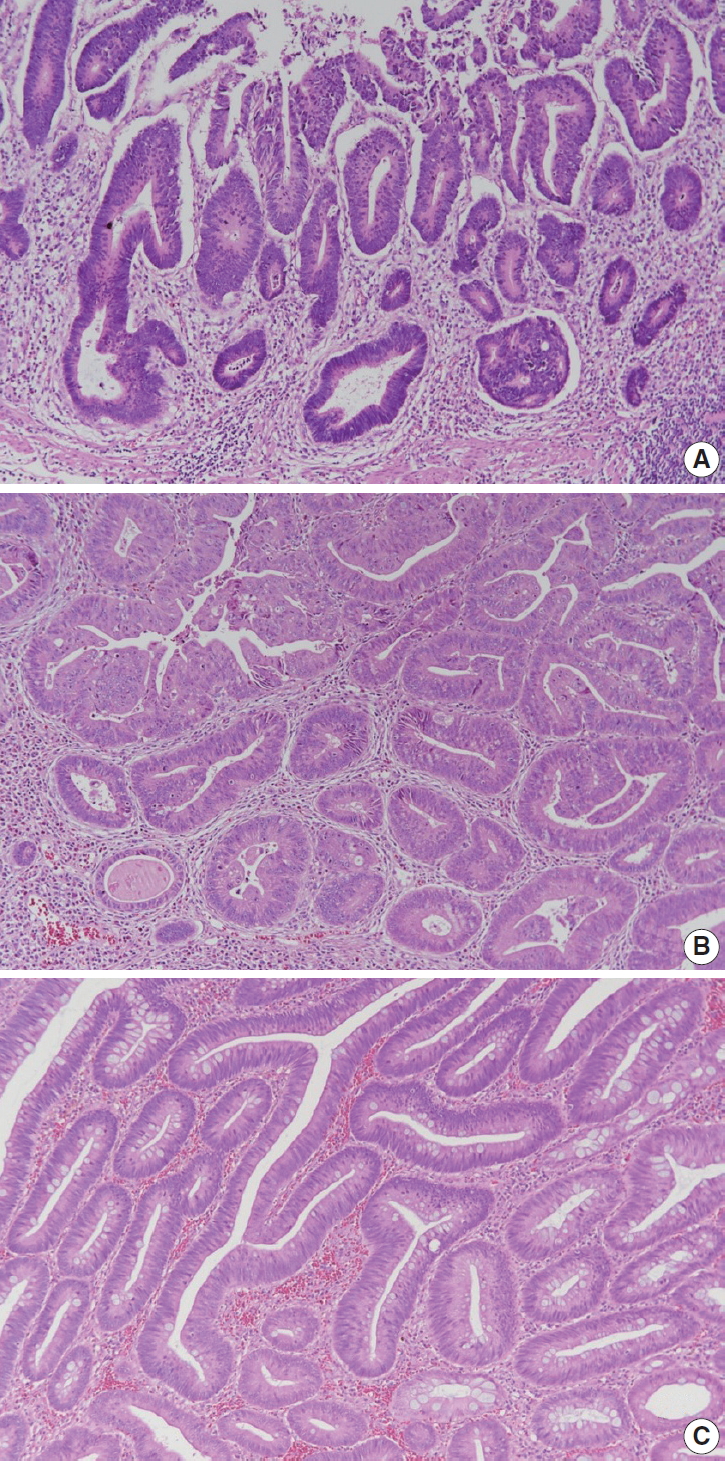
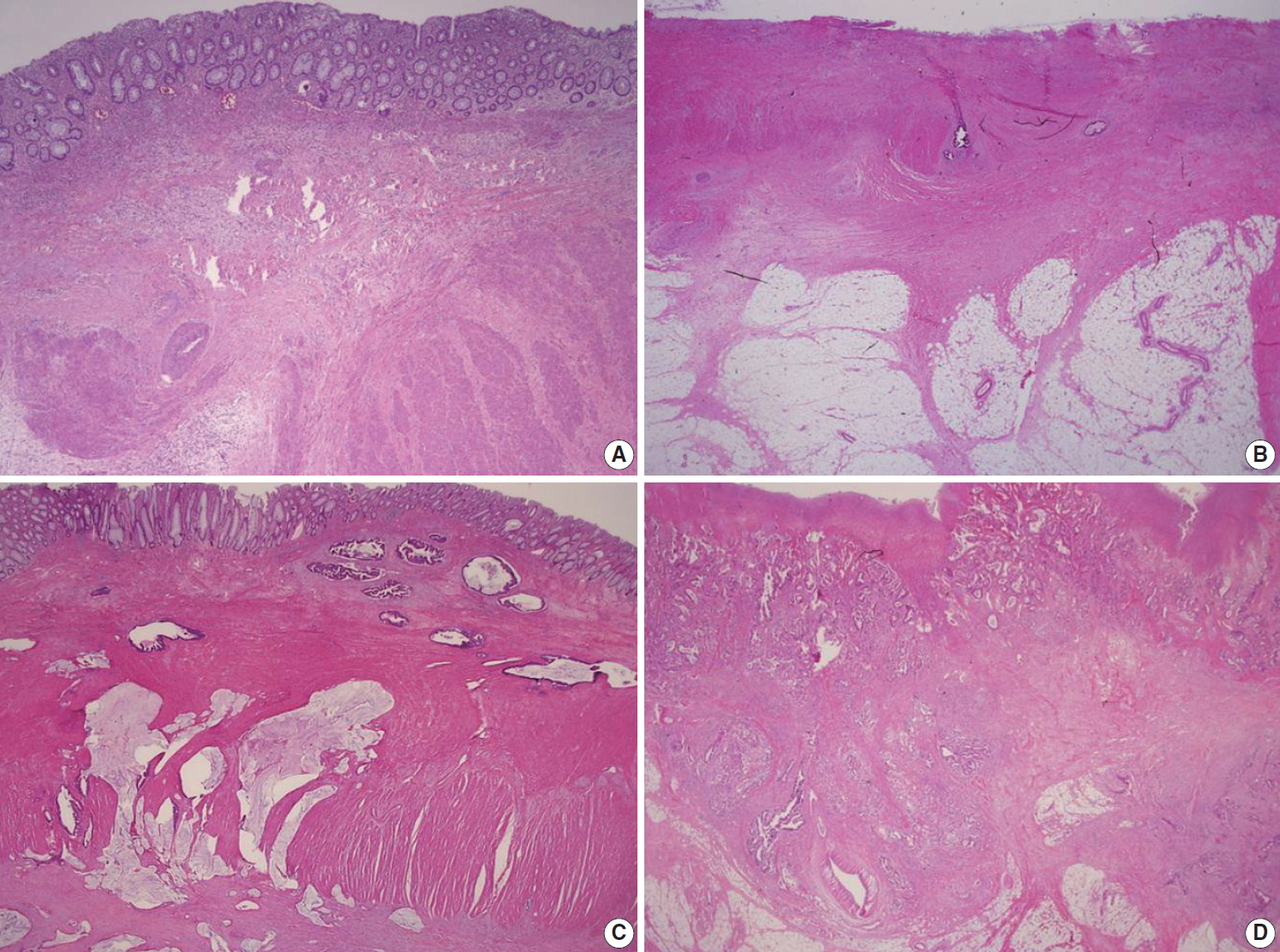
Fig. 3.
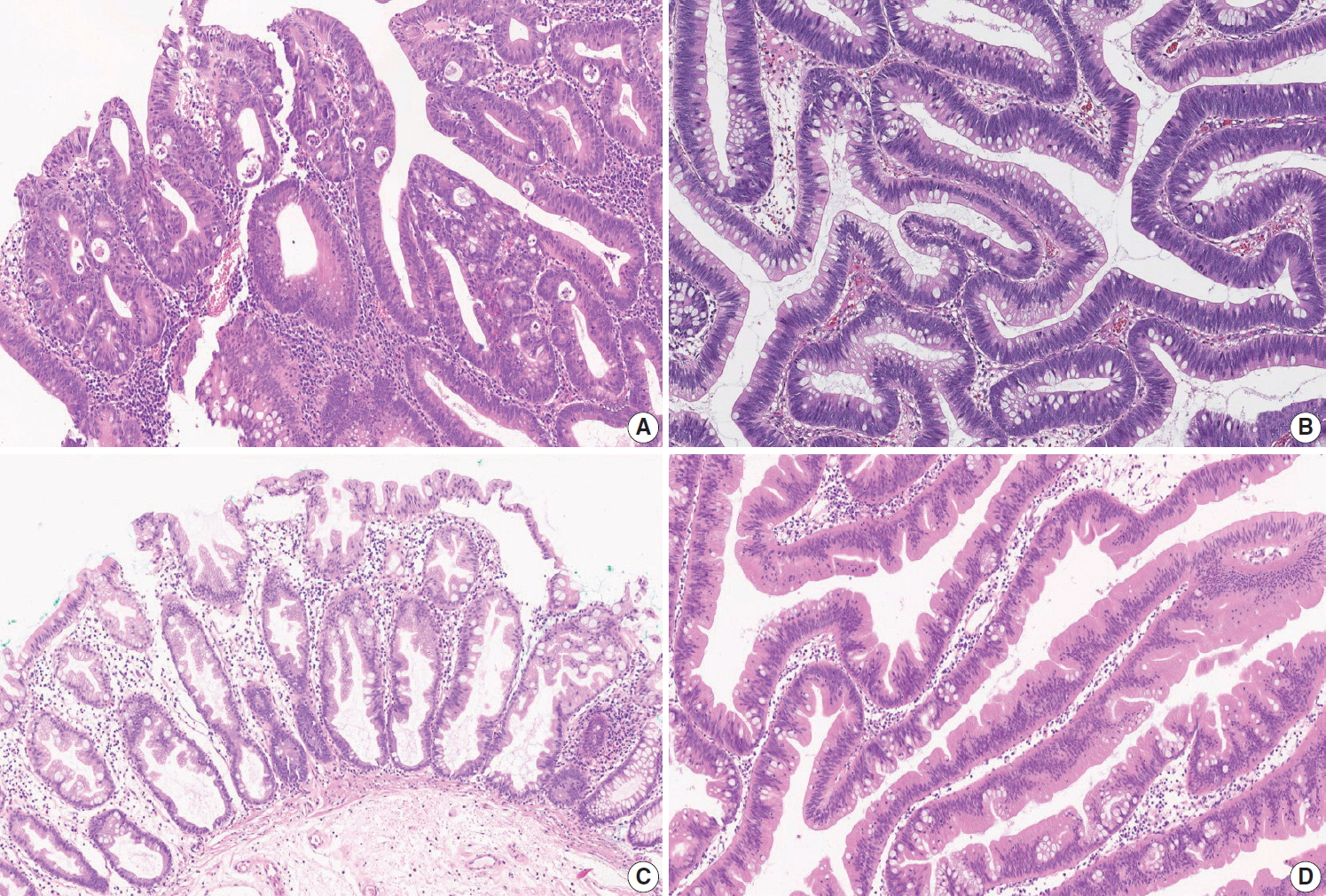
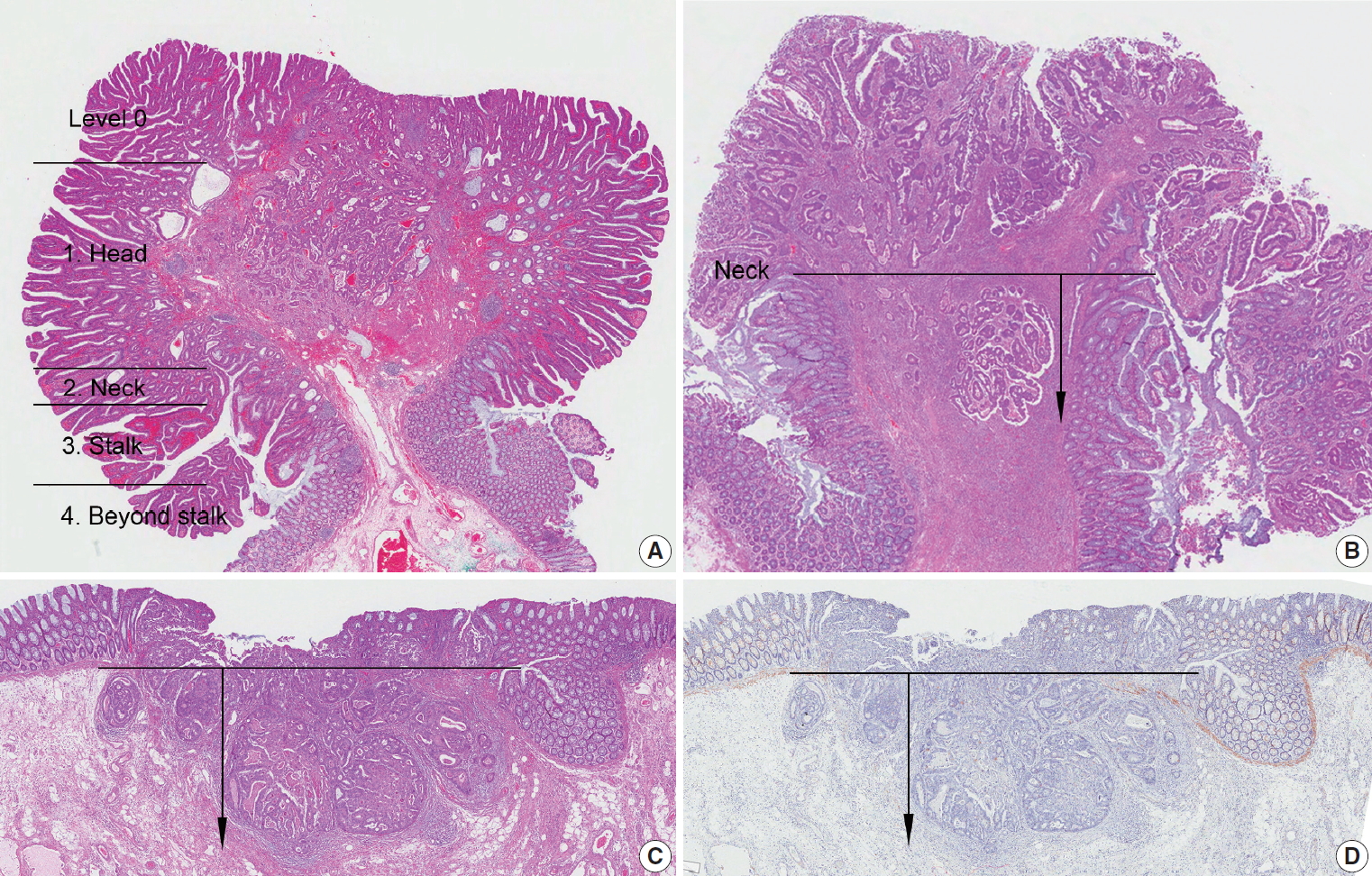
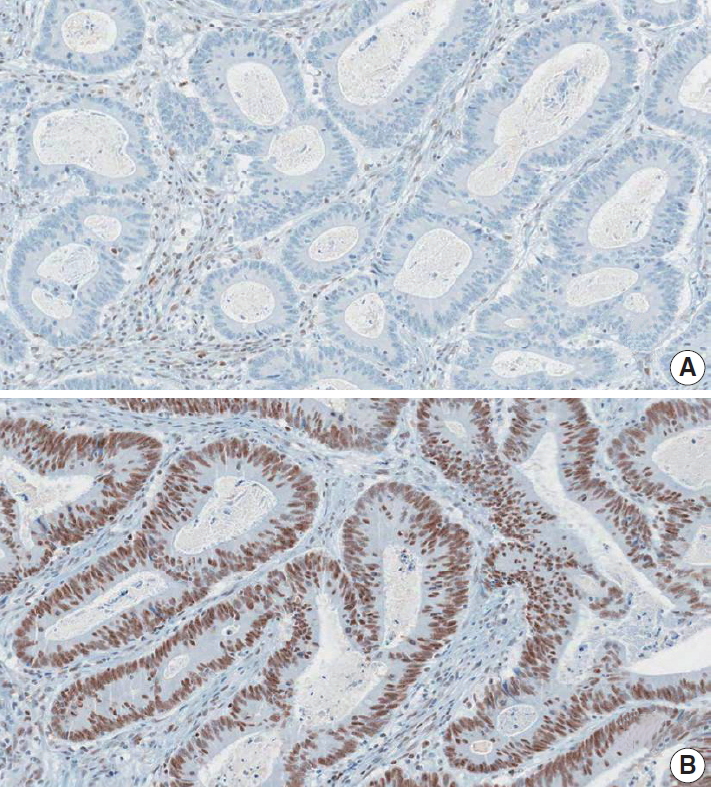
Fig. 4.
Fig. 5.
Fig. 6.
| Standard data elements | |||
|---|---|---|---|
| Specimen type | |||
| □ Right hemicolectomy | |||
| □ Transverse hemicolectomy | |||
| □ Left hemicolectomy | |||
| □ Anterior resection | |||
| □ Low anterior resection | |||
| □ Abdominoperineal resection | |||
| □ Subtotal/total colectomy | |||
| □ Total proctocolectomy | |||
| □ Transanal excision | |||
| □ Endoscopic mucosal resection | |||
| □ Other: (specify: ) | |||
| Histopathologic type of invasive carcinoma | |||
| □ Adenocarcinoma, NOS | |||
| □ Low-grade (well differentiated and moderately differentiated) | |||
| □ High-grade (poorly differentiated) | |||
| □ Mucinous adenocarcinoma | |||
| □ Signet ring cell carcinoma | |||
| □ Medullary carcinoma | |||
| □ Serrated adenocarcinoma | |||
| □ Micropapillary adenocarcinoma | |||
| □ Squamous cell (epidermoid) carcinoma (excluding upwardly spreading anal tumors) | |||
| □ Adenosquamous carcinoma | |||
| □ Small cell neuroendocrine carcinoma | |||
| □ Large cell neuroendocrine carcinoma | |||
| □ Mixed neuroendocrine-non-neuroendocrine neoplasm | |||
| □ Undifferentiated carcinoma | |||
| □ Other: (specify: ) | |||
| Location | |||
| □ Cecum | |||
| □ Ascending colon | |||
| □ Hepatic flexure | |||
| □ Transverse colon | |||
| □ Splenic flexure | |||
| □ Descending colon | |||
| □ Sigmoid colon | |||
| □ Rectosigmoid junction | |||
| □ Rectum | |||
| □ Other: (specify: ) | |||
| Gross type | |||
| □ Fungating/polypoid | |||
| □ Ulcerofungating | |||
| □ Ulceroinfiltrative | |||
| □ Infiltrative | |||
| □ Unclassifiable | |||
| Tumor size | |||
| × × cm | |||
| Depth of invasion | |||
| □ Intramucosal carcinoma (pTis) | |||
| □ Tumor invades the submucosa (pT1) | |||
| □ Tumor invades the muscularis propria (pT2) | |||
| □ Tumor invades through the muscularis propria into pericolorectal tissue (pT3) | |||
| □ Tumor invades through the visceral peritoneum (pT4a) | |||
| □ Tumor directly invades or adheres to adjacent organs or structures (pT4b) | |||
| - [Endoscopic excision (Endoscopic submucosal dissection/polypectomy) or transanal excision] | |||
| □ Tumor invades the lamina propria with no extension through muscularis mucosae (pTis) | |||
| □ Tumor invades the submucosa (pT1) | |||
| For sessile lesion: | |||
| Distance of tumor from muscularis mucosae: mm | |||
| For pedunculated lesion: | |||
| Haggitt level (head, neck, stalk, beyond stalk) | |||
| Distance of tumor invasion in stalk: mm | |||
| Resection margin | |||
| Proximal margin | |||
| □ Free from carcinoma | |||
| □ Involved by carcinoma | |||
| Distal margin | |||
| □ Free from carcinoma | |||
| □ Involved by carcinoma | |||
| Circumferential margin (rectum only) | |||
| □ Free from carcinoma | |||
| □ Involved by carcinoma | |||
| Safety margin: proximal cm, distal cm, circumferential cm | |||
| - [Endoscopic excision (Endoscopic mucosal resection/submucosal dissection/polypectomy) or transanal excision] | |||
| Deep margin | |||
| □ Free from carcinoma | |||
| □ Involved by carcinoma | |||
| □ Not applicable | |||
| Horizontal margin | |||
| □ Free from carcinoma | |||
| □ Involved by carcinoma | |||
| □ Involved by adenoma: (specify grade: ) | |||
| □ Not applicable | |||
| Safety margin: deep cm, horizontal cm | |||
| Regional lymph node metastasis | |||
| □ No metastasis in all regional lymph nodes (pN0) | |||
| □ Metastasis to out of regional lymph nodes pN | |||
| Lymphatic (small vessel) invasion | |||
| □ Not identified | |||
| □ Present | |||
| Venous invasion | |||
| □ Not identified | |||
| □ Present | |||
| □ Intramural | |||
| □ Extramural | |||
| Perineural invasion | |||
| □ Not identified | |||
| □ Present | |||
| Pre-existing adenoma | |||
| □ Absent | |||
| □ Tubular/Tubulovillous/Villous adenoma | |||
| Low grade dysplasia/High grade dysplasia | |||
| □ Sessile serrated lesion (sessile serrated adenoma/polyp) | |||
| □ Sessile serrated lesion (sessile serrated adenoma/polyp) with dysplasia | |||
| □ Traditional serrated adenoma | |||
| □ Other (specify: ) | |||
| Associated findings | |||
| □ Absent | |||
| □ Tumor perforation (pT4a) | |||
| □ Perforation (non-tumor perforation) | |||
| □ Metastasis to one site or organ without peritoneal metastasis (pM1a) | |||
| □ Metastasis to two or more sites or organs without peritoneal metastasis (pM1b) | |||
| □ Metastasis to the peritoneal surface with or without other site or organ metastasis (pM1c) | |||
| Specify metastatic sites or organs: | |||
| Separate lesions | |||
| □ Absent | |||
| □ Adenoma | |||
| □ Polyp | |||
| □ GIST | |||
| □ Ulcerative colitis/Crohn’s disease | |||
| □ Others | |||
| Specify: | |||
Conditional data elements |
|||
| Tumor budding | |||
| □ Not identified | |||
| □ Present | |||
| □ ≤ 4 buds (low) | |||
| □ 5–9 buds (intermediate) | |||
| □ ≥ 10 buds (high) | |||
| □ Cannot be assessed (specify: ) | |||
| Completeness of total mesorectal excision | |||
| □ Complete | |||
| □ Nearly complete | |||
| □ Incomplete | |||
| □ Cannot be determined | |||
| Preoperative chemoradiotherapy | |||
| □ Yes □ No □ Not known | |||
| If yes) Tumor regression grade | |||
| □ Grade 0: No viable cancer cells (complete response) | |||
| □ Grade 1: Single cells or rare small groups of cancer cells (near-complete response) | |||
| □ Grade 2: Residual cancer with evident tumor regression, but more than single cells or rare small groups of cancer cells (partial response) | |||
| □ Grade 3: Extensive residual cancer with no evident tumor regression (poor or no response) | |||
| DNA mismatch repair immunohistochemistry | |||
| MLH1: | □ Positive (retained expression) | ||
| □ Negative (loss of expression) | |||
| MSH2: | □ Positive (retained expression) | ||
| □ Negative (loss of expression) | |||
| PMS2: | □ Positive (retained expression) | ||
| □ Negative (loss of expression) | |||
| MSH6: | □ Positive (retained expression) | ||
| □ Negative (loss of expression) | |||
| Summary: DNA mismatch repair deficiency (was/was not) observed | |||
| Microsatellite instability (MSI) | |||
| Summary: | □ MSI-stable (MSS) | ||
| □ MSI-low (MSI-L) | |||
| □ MSI-high (MSI-H) | |||
| KRAS mutation analysis | |||
| □ No mutation detected | |||
| □ Mutation detected (specify: example: c.35G > A, p.Gly12Asp ) | |||
| NRAS mutation analysis | |||
| □ No mutation detected | |||
| □ Mutation detected (specify: example: c.35G>A, p.Gly12Asp ) | |||
| BRAF mutation analysis | |||
| □ No mutation detected | |||
| □ BRAF V600E (c.1799T > A) mutation | |||
| □ Other BRAF mutation (specify: ) | |||
Comment: This report is intended to be applicable to endoscopic resection or transanal excision specimens as well as surgical resection of CRC. NOS, not otherwise specified; CRC, colorectal cancer.

 E-submission
E-submission